Shows
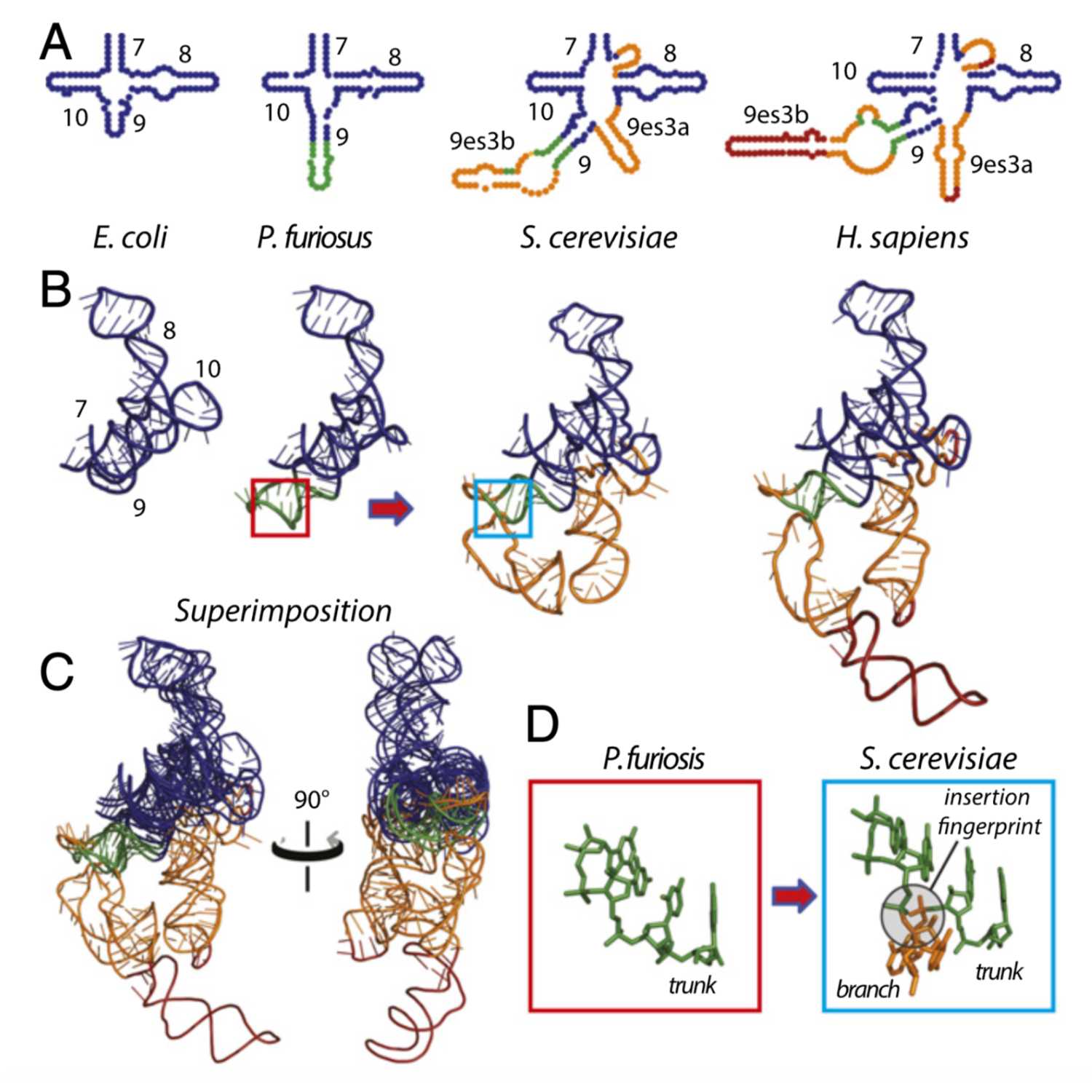
Papers Aloud#44 - History of the ribosome and the origin of translationTitle: History of the ribosome and the origin of translationAuthors: Anton S. Petrova, Burak Gulen, Ashlyn M. Norris, Nicholas A. Kovacs, Chad R. Bernier, Kathryn A. Lanier, George E. Fox, Stephen C. Harvey, Roger M. Wartell, Nicholas V. Hud, and Loren Dean Williams,Citation: Petrov, A. S., Gulen, B., Norris, A. M., Kovacs, N. A., Bernier, C. R., Lanier, K. A., Fox, G. E., Harvey, S. C., Wartell, R. M., Hud, N. V., & Williams, L. D. (2015). History of the ribosome and the origin of translation. Proceedings of the National Academy of Sciences of the United States of America, 112(50), 15396–15401.Link to ar...
2023-04-1110 min
Papers Aloud#44 - Histone-organized chromatin in bacteria - Hocher et al 2023 - BioRxivTitle: Histone-organized chromatin in bacteriaAuthors: Antoine Hocher, Shawn P. Laursen, Paul Radford, Jess Tyson, Carey Lambert, Kathryn M Stevens, Mathieu Picardeau, R. Elizabeth Sockett, Karolin Luger, Tobias WarneckeCitation: Hocher, A., Laursen, S. P., Radford, P., Tyson, J., Lambert, C., Stevens, K. M., Picardeau, M., Elizabeth Sockett, R., Luger, K., & Warnecke, T. (2023). Histone-organized chromatin in bacteria. In bioRxiv (p. 2023.01.26.525422). https://doi.org/10.1101/2023.01.26.525422Link to paper
2023-02-2210 min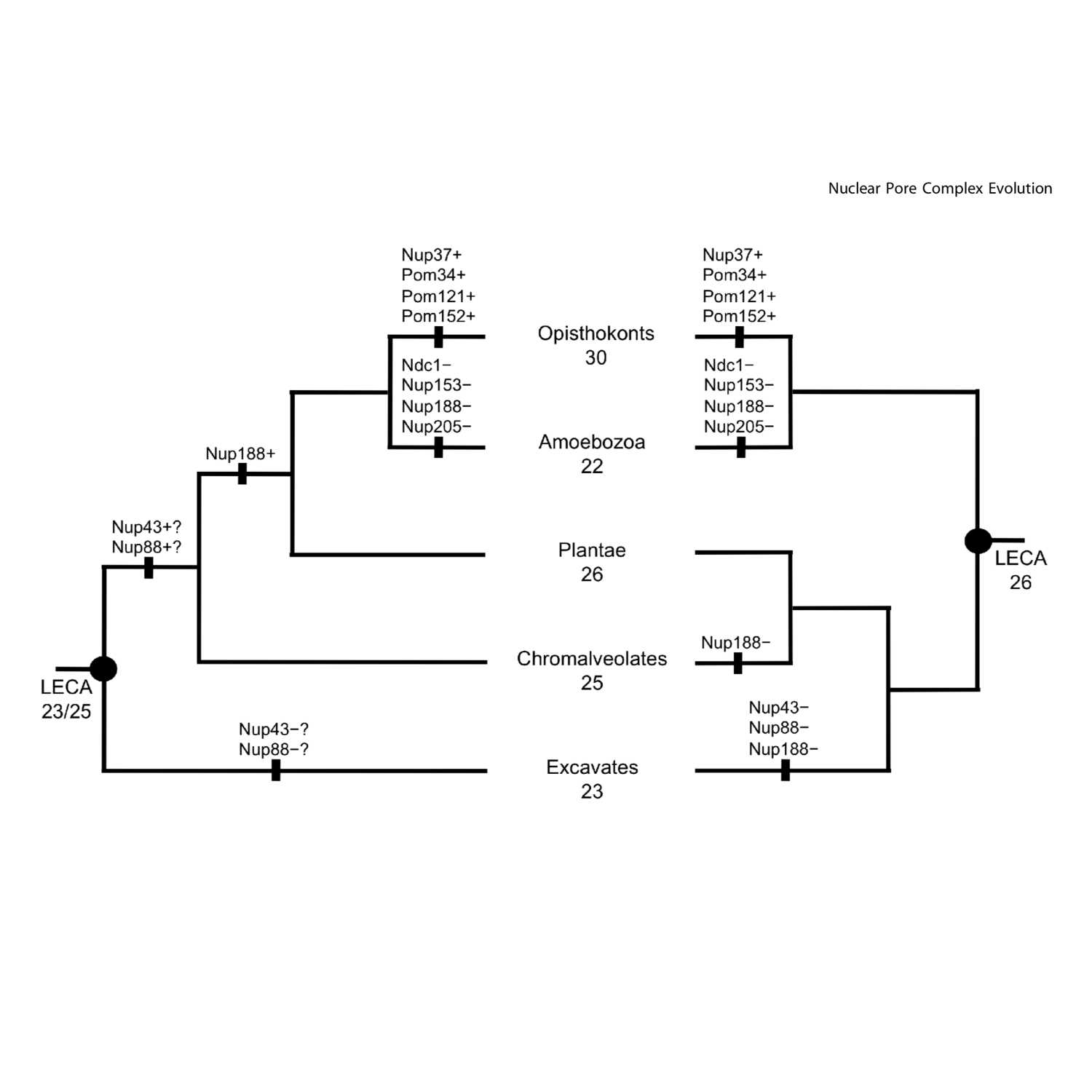
Papers Aloud#44 - A Complete Nuclear Pore Complex in the Last Eukaryotic Common Ancestor - Neumann et al., 2010 - PLON OneTitle: Comparative Genomic Evidence for a Complete Nuclear Pore Complex in the Last Eukaryotic Common AncestorAuthors: Nadja Neumann, Daniel Lundin, Anthony M. PooleCitation: Neumann, N., Lundin, D., & Poole, A. M. (2010). Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PloS One, 5(10), e13241.Link to paper
2023-02-1535 min
Papers Aloud#44 - Origin of the nucleus - Jékely 2008 - Biology DirectTitle: Origin of the nucleus and Ran-dependent transport to safeguard ribosome biogenesis in a chimeric cellAuthors: Gáspár JékelyCitation: Jékely, G. (2008). Origin of the nucleus and Ran-dependent transport to safeguard ribosome biogenesis in a chimeric cell. Biology Direct, 3, 31.Link to article
2023-02-0827 min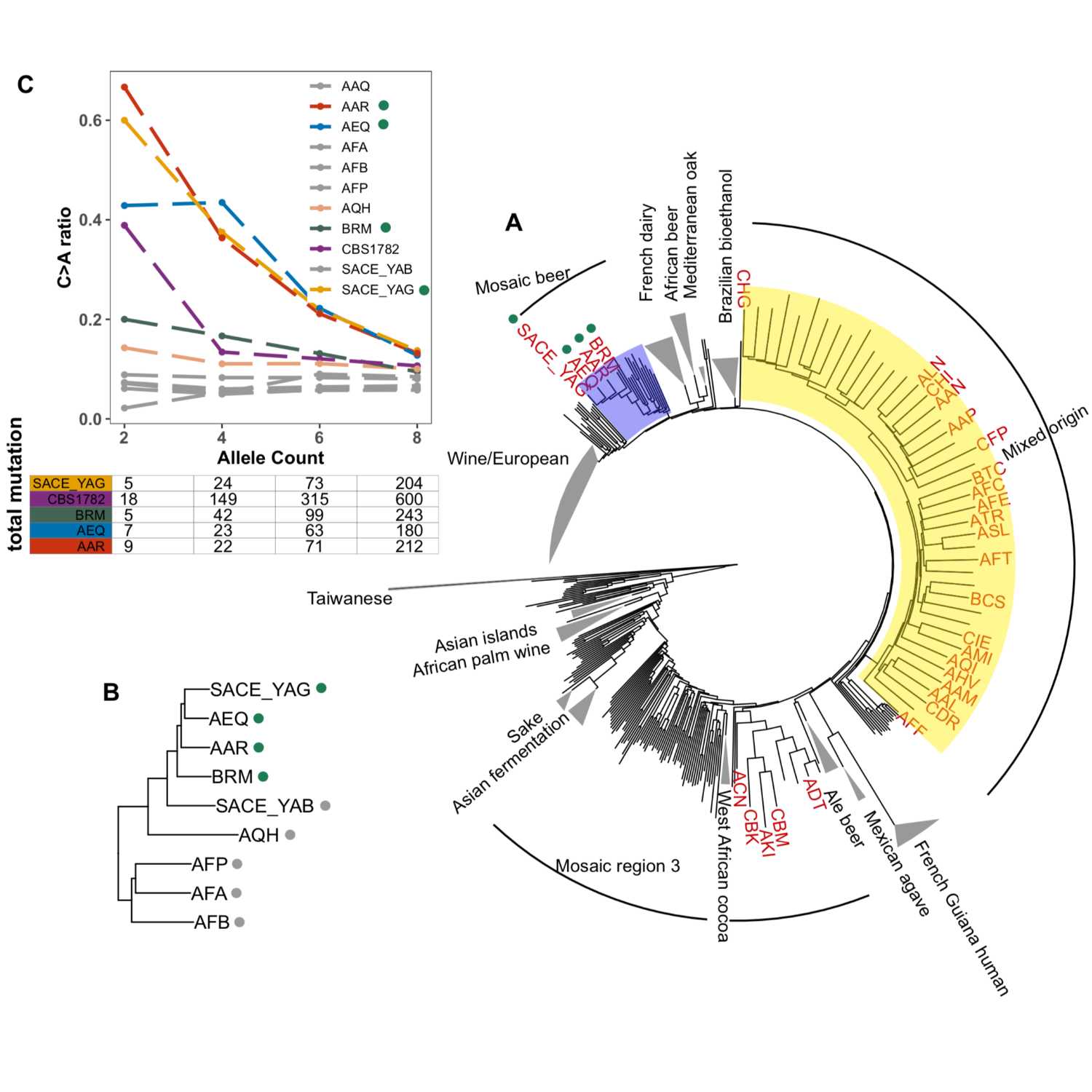
Papers Aloud#43 - Mutation Spectrum Variation - Jiang et al., 2021 - eLifeTitle: A modified fluctuation assay reveals a natural mutator phenotype that drives mutation spectrum variation within Saccharomyces cerevisiaeAuthors: Pengyao Jiang, Anja R Ollodart, Vidha Sudhesh, Alan J Herr, Maitreya J Dunham, Kelley HarrisCitation: Jiang, P., Ollodart, A. R., Sudhesh, V., Herr, A. J., Dunham, M. J., & Harris, K. (2021). A modified fluctuation assay reveals a natural mutator phenotype that drives mutation spectrum variation within Saccharomyces cerevisiae. eLife, 10. https://doi.org/10.7554/eLife.68285.Link to article
2023-02-0626 min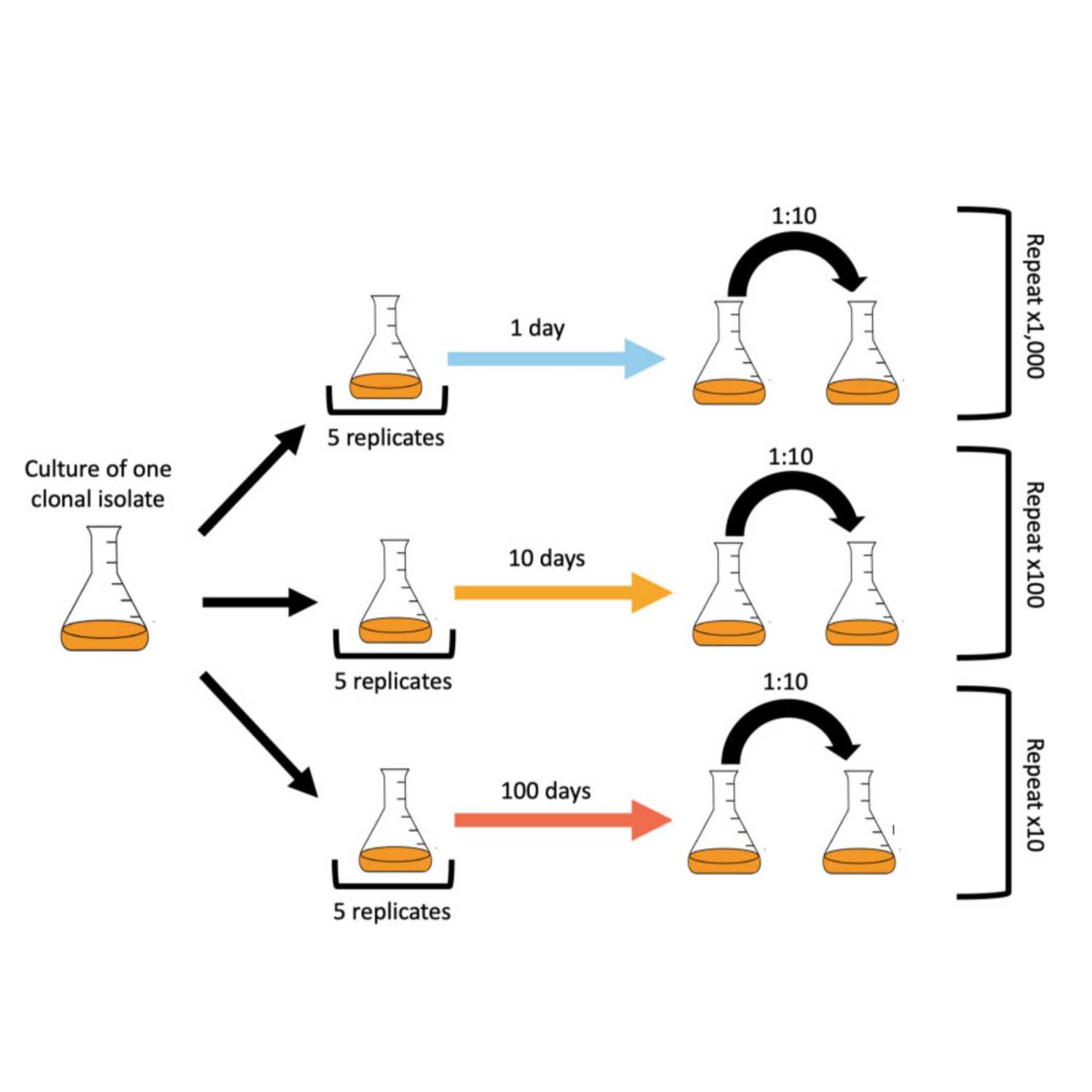
Papers Aloud#42 - Evolution in Energy Limited Microorganisms - Shoemaker et al., 2021 - MBETitle: Molecular Evolutionary Dynamics of Energy Limited MicroorganismsAuthors: William R. Shoemaker, Evgeniya Polezhaeva, Kenzie B. Givens, and Jay T. LennonCitation: Shoemaker, W. R., Polezhaeva, E., Givens, K. B., & Lennon, J. T. (2021). Molecular Evolutionary Dynamics of Energy Limited Microorganisms. Molecular Biology and Evolution, 38(10), 4532–4545.Link to article
2023-02-0412 min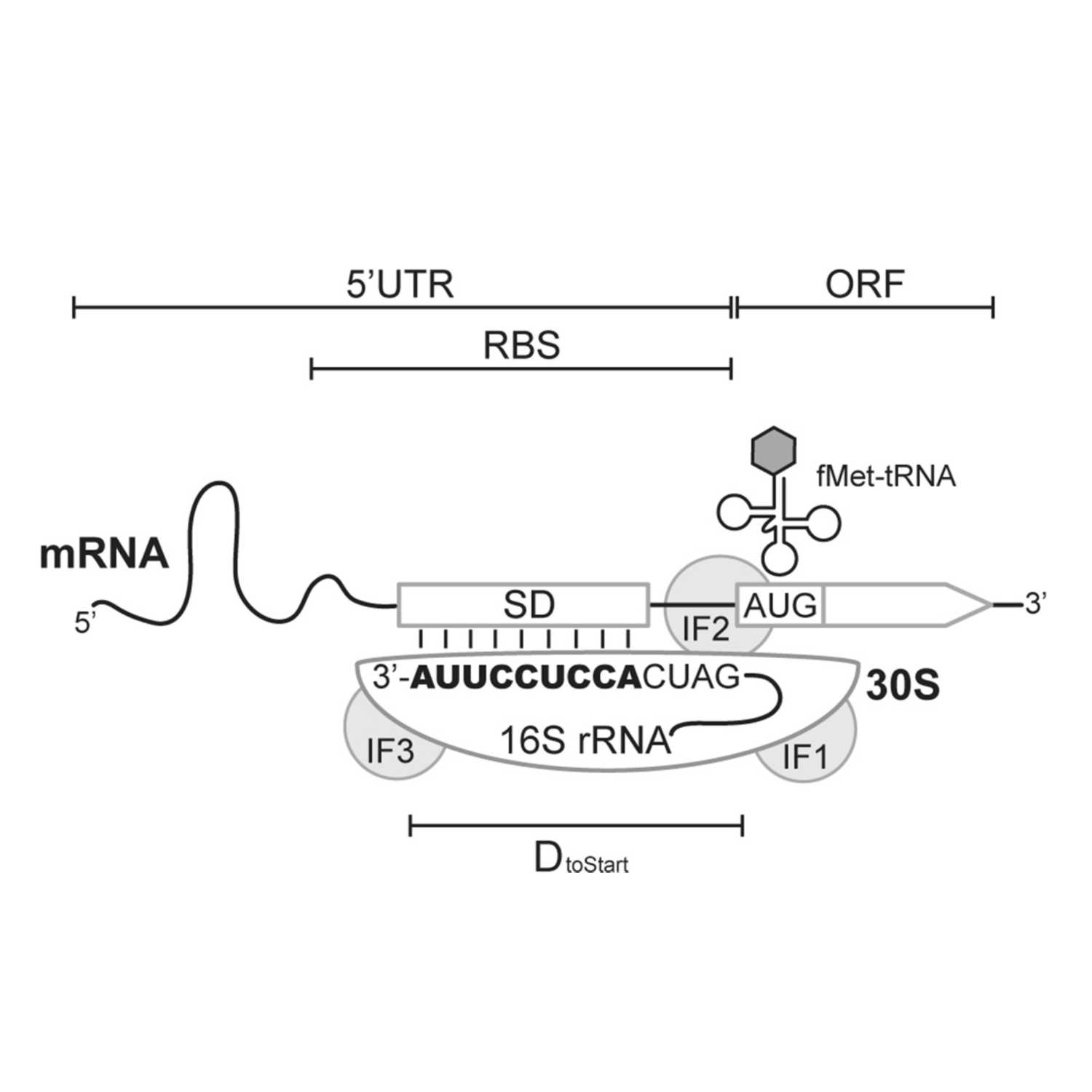
Papers Aloud#41 - Diversity of Shine-Dalgarno Sequences - Wen et al., 2021 - RNA BiologyTitle: The diversity of Shine-Dalgarno sequences sheds light on the evolution of translation initiationAuthors: Jin-Der Wen, Syue-Ting Kuo, and Hsin-Hung David Chou.Citation: Wen, J.-D., Kuo, S.-T., & Chou, H.-H. D. (2021). The diversity of Shine-Dalgarno sequences sheds light on the evolution of translation initiation. RNA Biology, 18(11), 1489–1500.Link to article
2023-01-2647 min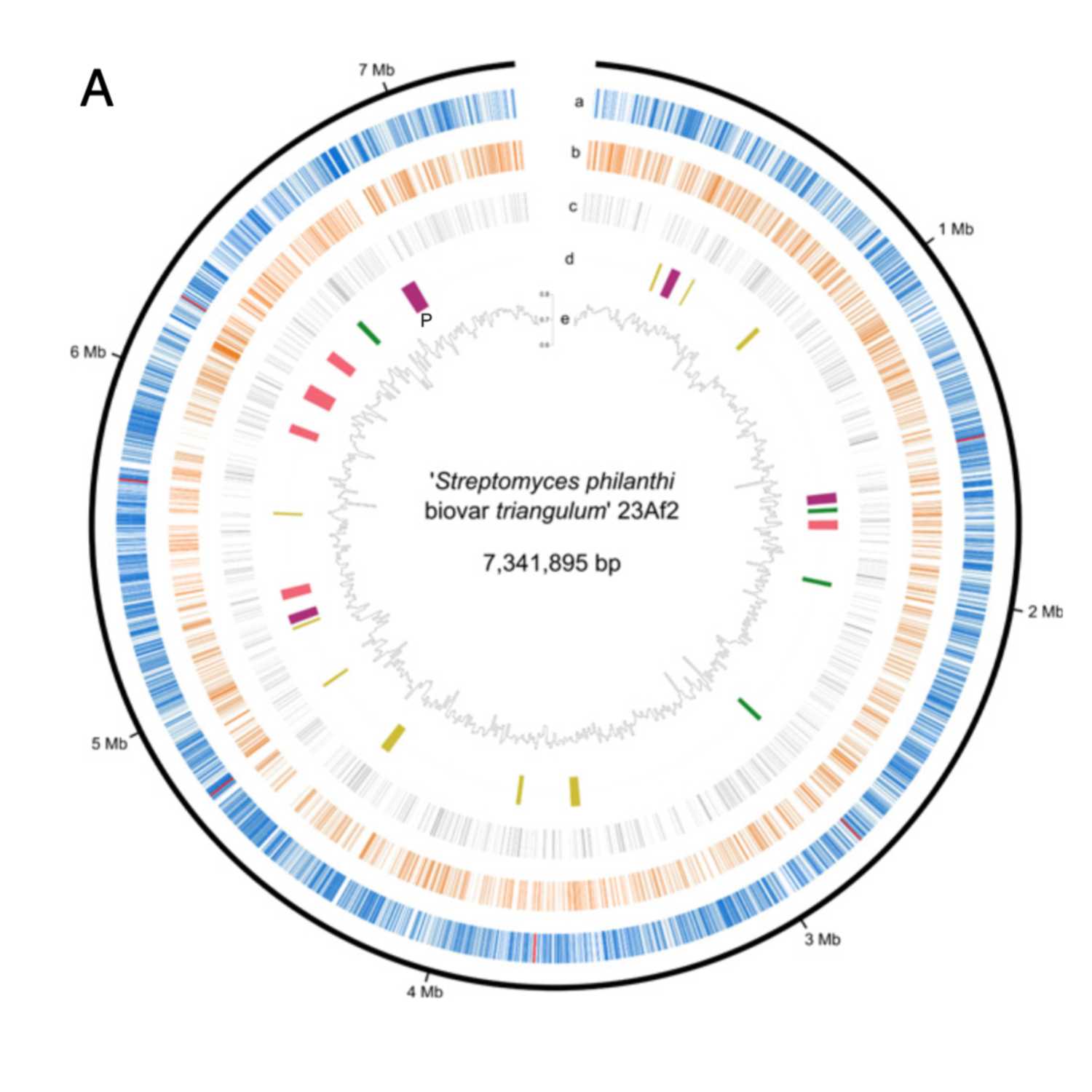
Papers Aloud#40 - Incipient genome erosion and metabolic streamlining for antibiotic production in a defensive symbiont - Nechitaylo et al., 2022 - PNASTitle: Incipient genome erosion and metabolic streamlining for antibiotic production in a defensive symbiontAuthors: Taras Y. Nechitaylo, Mario Sandoval-Calderónb\, Tobias Engl, Natalie Wielsch, Diane M. Dunne, Alexander Goesmann, Erhard Strohm, Aleš Svatoš, Colin Dale, Robert B. Weiss, and Martin KaltenpothCitation: Nechitaylo, T. Y., Sandoval-Calderón, M., Engl, T., Wielsch, N., Dunn, D. M., Goesmann, A., Strohm, E., Svatoš, A., Dale, C., Weiss, R. B., & Kaltenpoth, M. (2021). Incipient genome erosion and metabolic streamlining for antibiotic production in a defensive symbiont. Proceedings of the National Academy of Sciences of the United States of America, 118(17). https...
2022-09-2333 min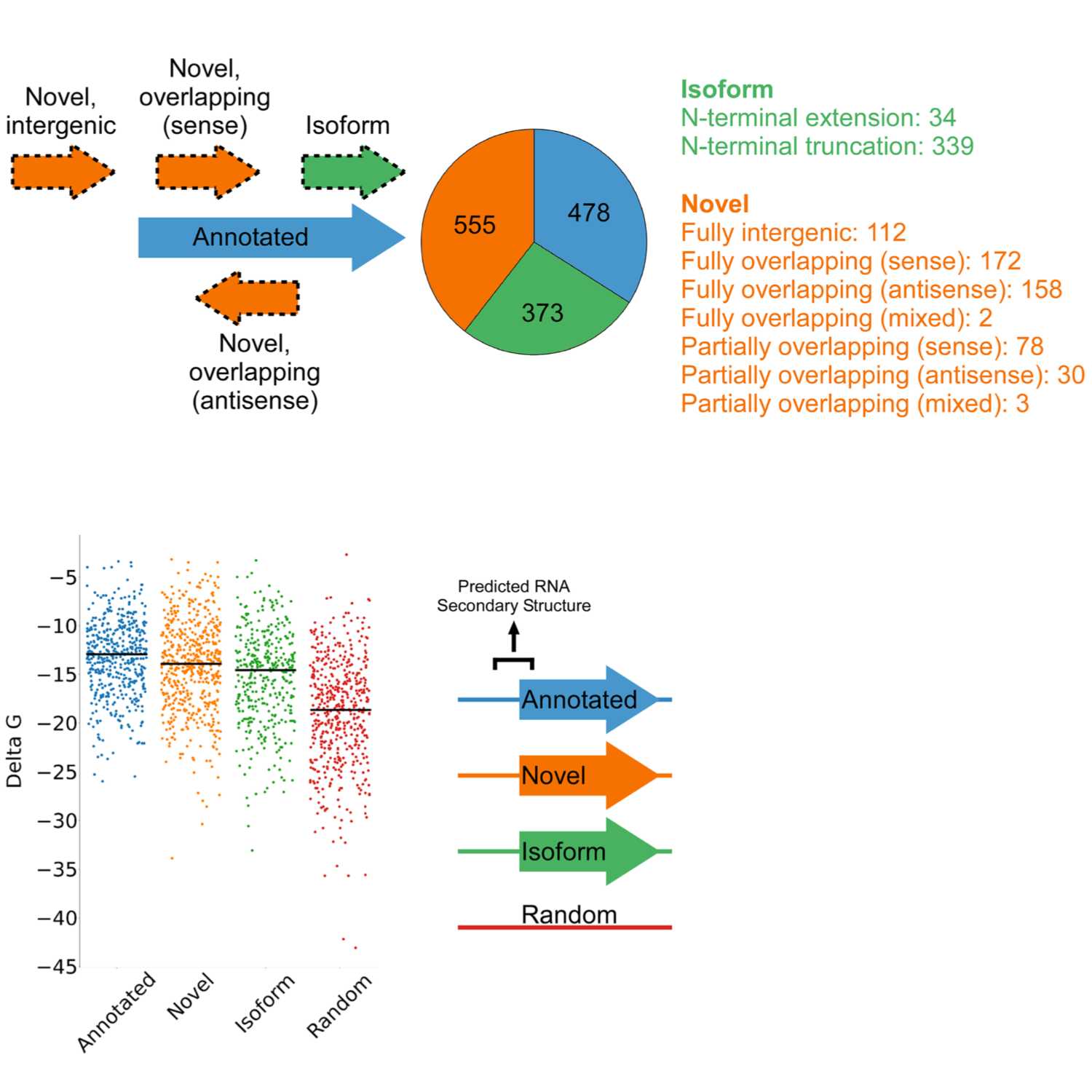
Papers Aloud#39 - Pervasive translation in Mycobacterium tuberculosis - Smith et al., 2022 - eLifeTitle: Pervasive translation in Mycobacterium tuberculosisAuthors: Carol Smith, Jill G Canestrari, Archer J Wang, Matthew M Champion, Keith M Derbyshire, Todd A Gray, Joseph T Wade.Citation: Smith, C., Canestrari, J. G., Wang, A. J., Champion, M. M., Derbyshire, K. M., Gray, T. A., & Wade, J. T. (2022). Pervasive translation in Mycobacterium tuberculosis. eLife, 11. https://doi.org/10.7554/eLife.73980Link to article
2022-06-0521 min
Papers Aloud# 38Simonsen, A. K. (2021). Environmental stress leads to genome streamlining in a widely distributed species of soil bacteria. The ISME Journal. https://doi.org/10.1038/s41396-021-01082-x
2021-10-1018 min
Papers Aloud# 38 - Environmental stress and genome streamlining - Simonsen, 2021 - ISMETitle: Environmental stress leads to genome streamlining in a widelydistributed species of soil bacteriaAuthors: Anna K. SimonsenCitation: Simonsen, A. K. (2021). Environmental stress leads to genome streamlining in a widely distributed species of soil bacteria. The ISME Journal. https://doi.org/10.1038/s41396-021-01082-xLink to article
2021-10-1018 min
Papers Aloud#38 - Are aliens among us - Davies, 2006 - Scientific AmericanTitle: Are Aliens among UsAuthors: Paul DaviesCitation: Davies, P. (2007). Are Aliens among Us? Scientific American, 297(6), 62–69.Link to article
2021-10-1027 min
Papers Aloud#37 - Environmental stress and genome streamlining - Simonsen, 2021 - ISMETitle: Environmental stress leads to genome streamlining in a widelydistributed species of soil bacteriaAuthors: Anna K. SimonsenCitation: Simonsen, A. K. (2021). Environmental stress leads to genome streamlining in a widely distributed species of soil bacteria. The ISME Journal. https://doi.org/10.1038/s41396-021-01082-xLink to article
2021-10-1018 min
Papers Aloud# 36 - Principles of Geobiochemistry - Shock and Boyd, 2015 - ElementsTitle: Principles of GeobiochemistryAuthors: Everett L. Shock and Eric S. BoydCitation: Shock, E. L., & Boyd, E. S. (2015). Principles of Geobiochemistry. Elements , 11(6), 395–401.Link to article
2021-09-2340 min
Papers Aloud#35 - The imitation game - Cronin et al., 2006 - Nature BiotechnologyTitle: The imitation game—a computational chemical approach to recognizing lifeAuthors: Leroy Cronin, Natalio Krasnogor, Benjamin G Davis, Cameron Alexander, Neil Robertson, Joachim H G Steinke, Sven L M Schroeder, Andrei N Khlobystov, Geoff Cooper, Paul M Gardner, Peter Siepmann, Benjamin J Whitaker & Dan MarshCitation: Cronin, L., Krasnogor, N., Davis, B. G., Alexander, C., Robertson, N., Steinke, J. H. G., Schroeder, S. L. M., Khlobystov, A. N., Cooper, G., Gardner, P. M., Siepmann, P., Whitaker, B. J., & Marsh, D. (2006). The imitation game--a computational chemical approach to recognizing life. Nature Biotechnology, 24(10), 1203–1206.Link to a...
2021-09-2317 min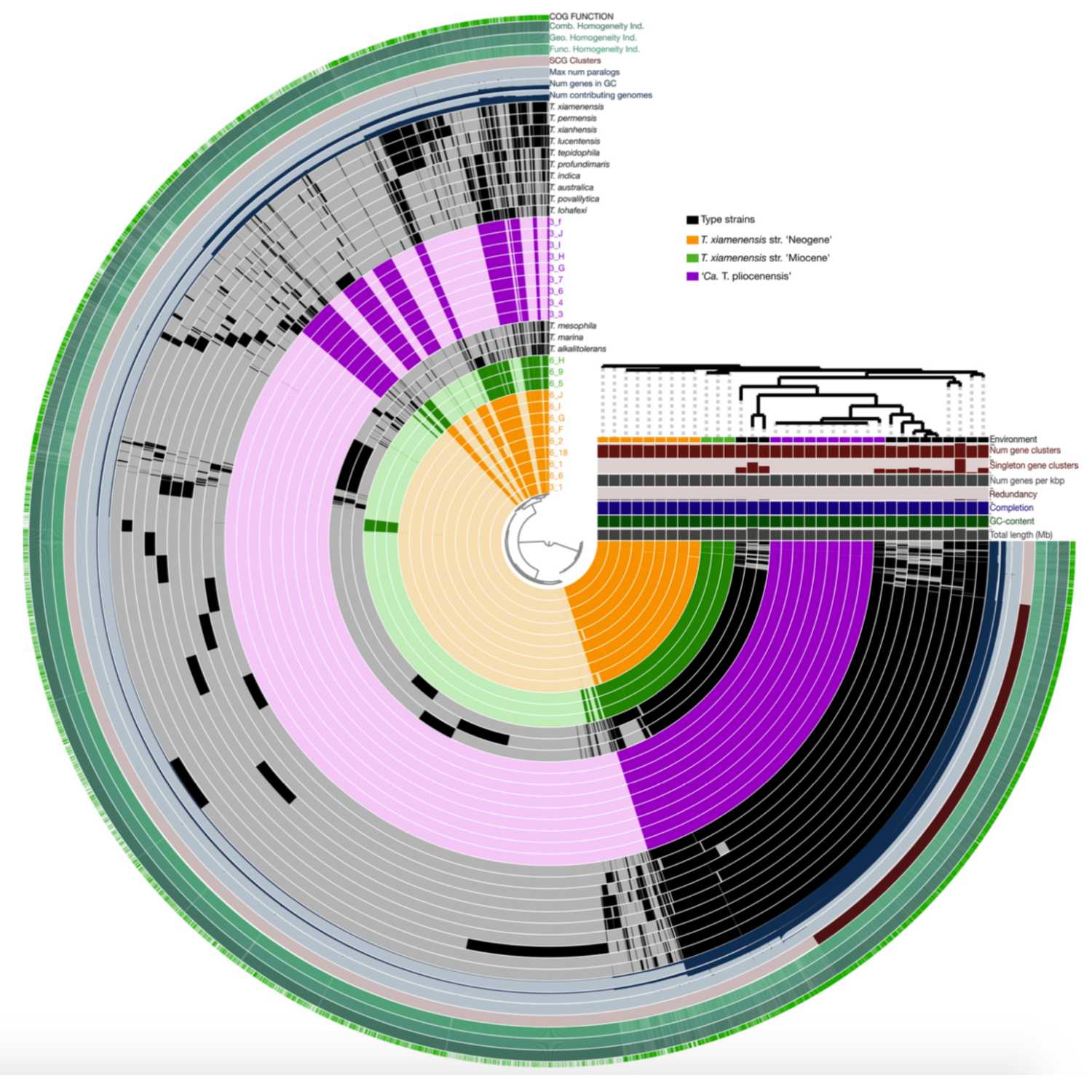
Papers Aloud#34 - Genome Evolution in Million-Year-Old Bacteria - Orsi et al., 2021 - mBioTitle: Genome Evolution in Bacteria Isolated from Million-Year-Old Subseafloor SedimentAuthors: William D. Orsi, Tobias Magritsch, Sergio Vargas, Ömer K. Coskun, Aurele Vuillemin, Sebastian Höhna, Gert Wörheide, Steven D’Hondt, B. Jesse Shapiro, Paul CariniCitation: Orsi, W. D., Magritsch, T., Vargas, S., Coskun, Ö. K., Vuillemin, A., Höhna, S., Wörheide, G., D’Hondt, S., Shapiro, B. J., & Carini, P. (2021). Genome Evolution in Bacteria Isolated from Million-Year-Old Subseafloor Sediment. mBio, 12(4), e0115021.Link to article
2021-09-0829 min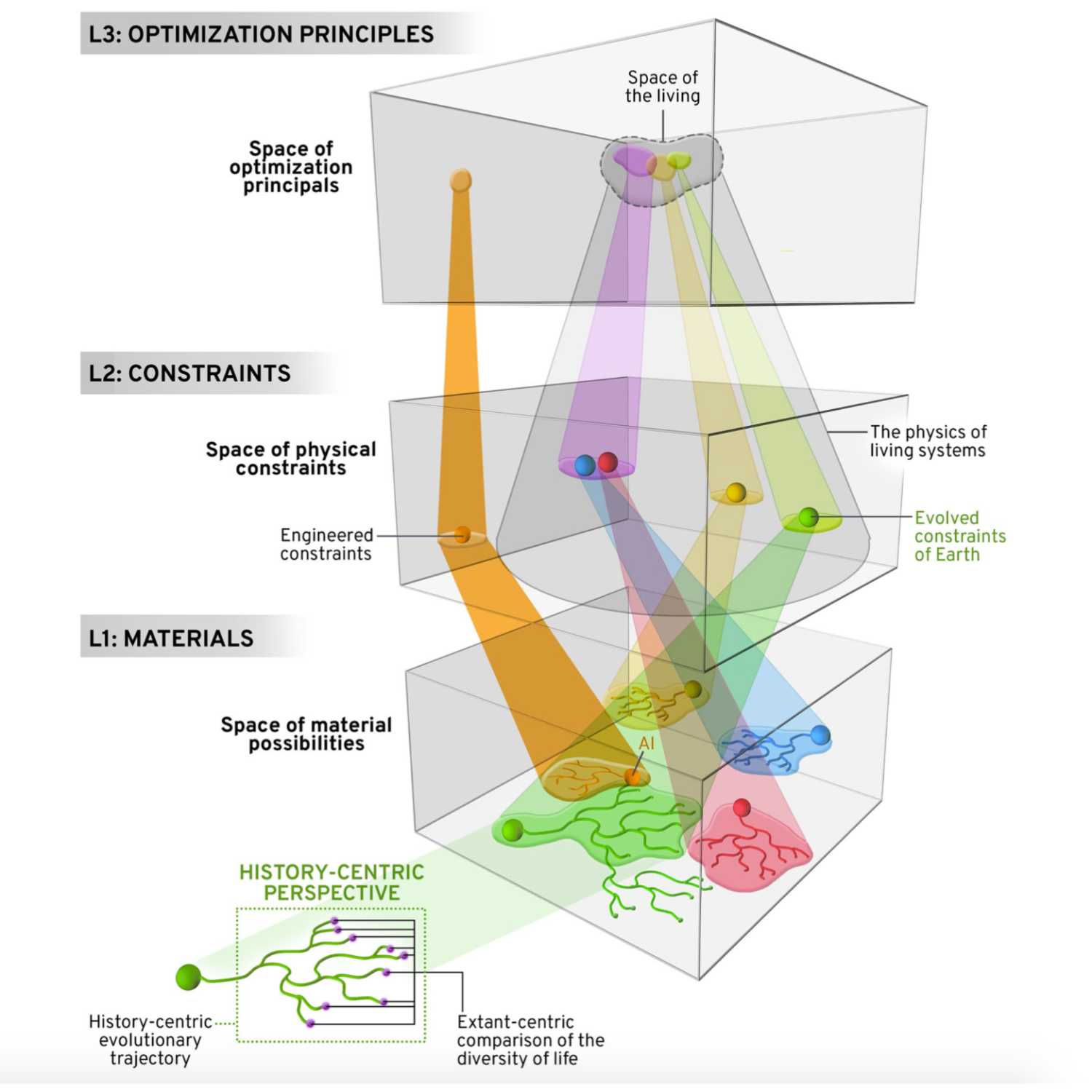
Papers Aloud#33 - Multiple Paths to Multiple Life - Kempes and Krakauer 2021 - Journal of Molecular EvolutionTitle: The Multiple Paths to Multiple LifeAuthors: Christopher P. Kempes and David C. KrakauerCitation: Kempes, C. P., & Krakauer, D. C. (2021). The Multiple Paths to Multiple Life. Journal of Molecular Evolution, 89(7), 415–426.Link to article
2021-09-0856 min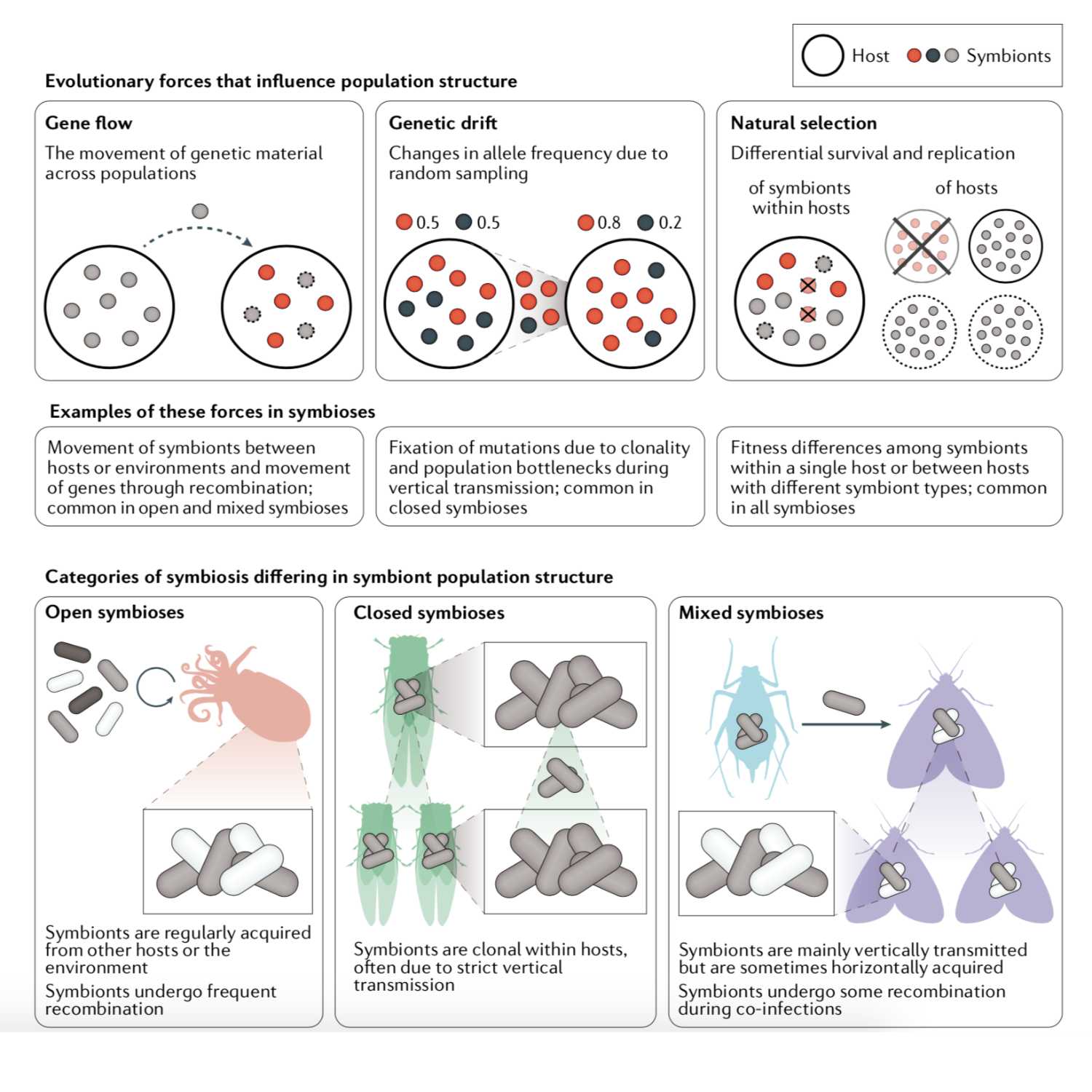
Papers Aloud#32 - Animal–microbe symbioses - Perreau and Moran, 2021 - Nature ReviewsTitle: Genetic innovations in animal–microbe symbiosesAuthors: Julie Perreau and Nancy A. MoranCitation: Perreau, J., & Moran, N. A. (2021). Genetic innovations in animal-microbe symbioses. Nature Reviews. Genetics. https://doi.org/10.1038/s41576-021-00395-zLink to article
2021-09-031h 10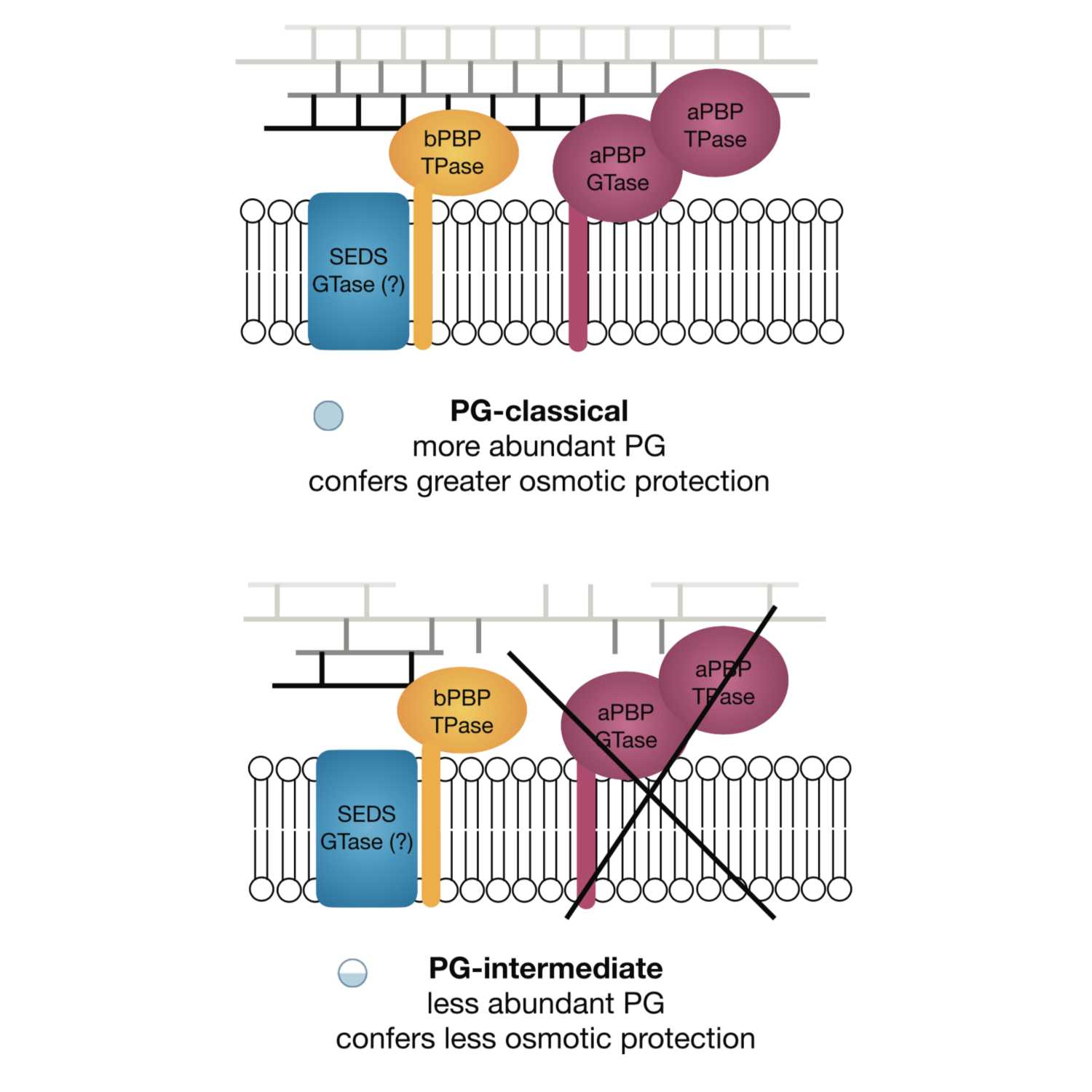
Papers Aloud#31 - Peptidoglycan-intermediate bacteria - Atwal et al., 2021 - mBioTitle: Discovery of a Diverse Set of Bacteria That Build Their Cell Walls without the Canonical Peptidoglycan Polymerase aPBP.Authors: Sharanjeet Atwal, Suthida Chuenklin, Edward M. Bonder, Juan Flores, Joseph J. Gillespie, Timothy P. Driscoll, Jeanne Salje.Citation: Atwal, S., Chuenklin, S., Bonder, E. M., Flores, J., Gillespie, J. J., Driscoll, T. P., & Salje, J. (n.d.). Discovery of a Diverse Set of Bacteria That Build Their Cell Walls without the Canonical Peptidoglycan Polymerase aPBP. https://doi.org/10.1128/mBio.01342-21.Link to article
2021-08-1613 min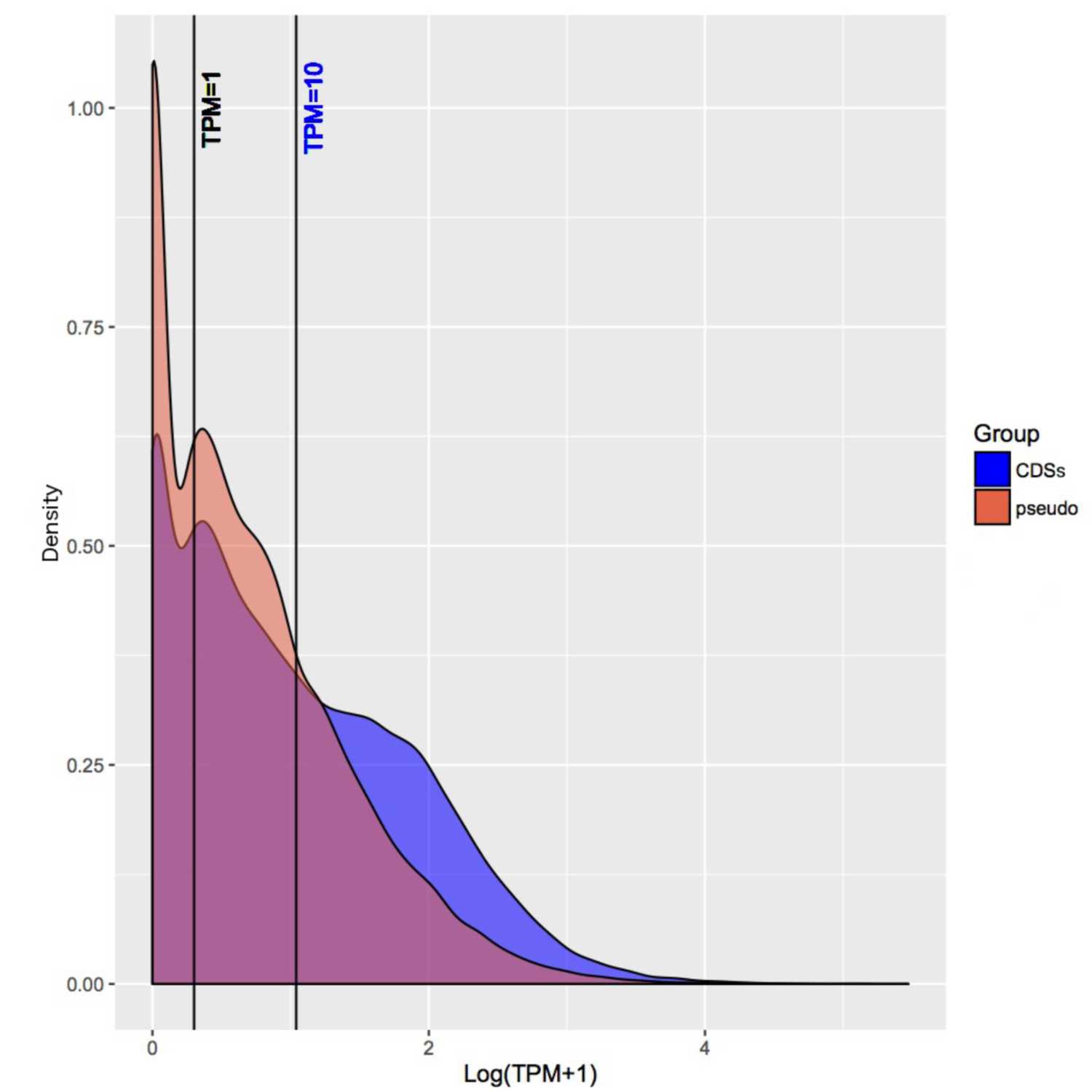
Papers Aloud#30 - Pseudogenes and gene expression - Goodhead et al., 2020 - Microbial GenomicsTitle: Large-scale and significant expression from pseudogenes in Sodalis glossinidius – a facultative bacterial endosymbiont. Authors: Ian Goodhead, Frances Blow, Philip Brownridge, Margaret Hughes, John Kenny, Ritesh Krishna, Lynn McLean, Pisut Pongchaikul, Rob Beynon, and Alistair C. Darby. Citation: Goodhead, I., Blow, F., Brownridge, P., Hughes, M., Kenny, J., Krishna, R., McLean, L., Pongchaikul, P., Beynon, R., & Darby, A. C. (2020). Large-scale and significant expression from pseudogenes in Sodalis glossinidius - a facultative bacterial endosymbiont. Microbial Genomics, 6(1). https://doi.org/10.1099/mgen.0.000285. Link to article
2021-08-1327 min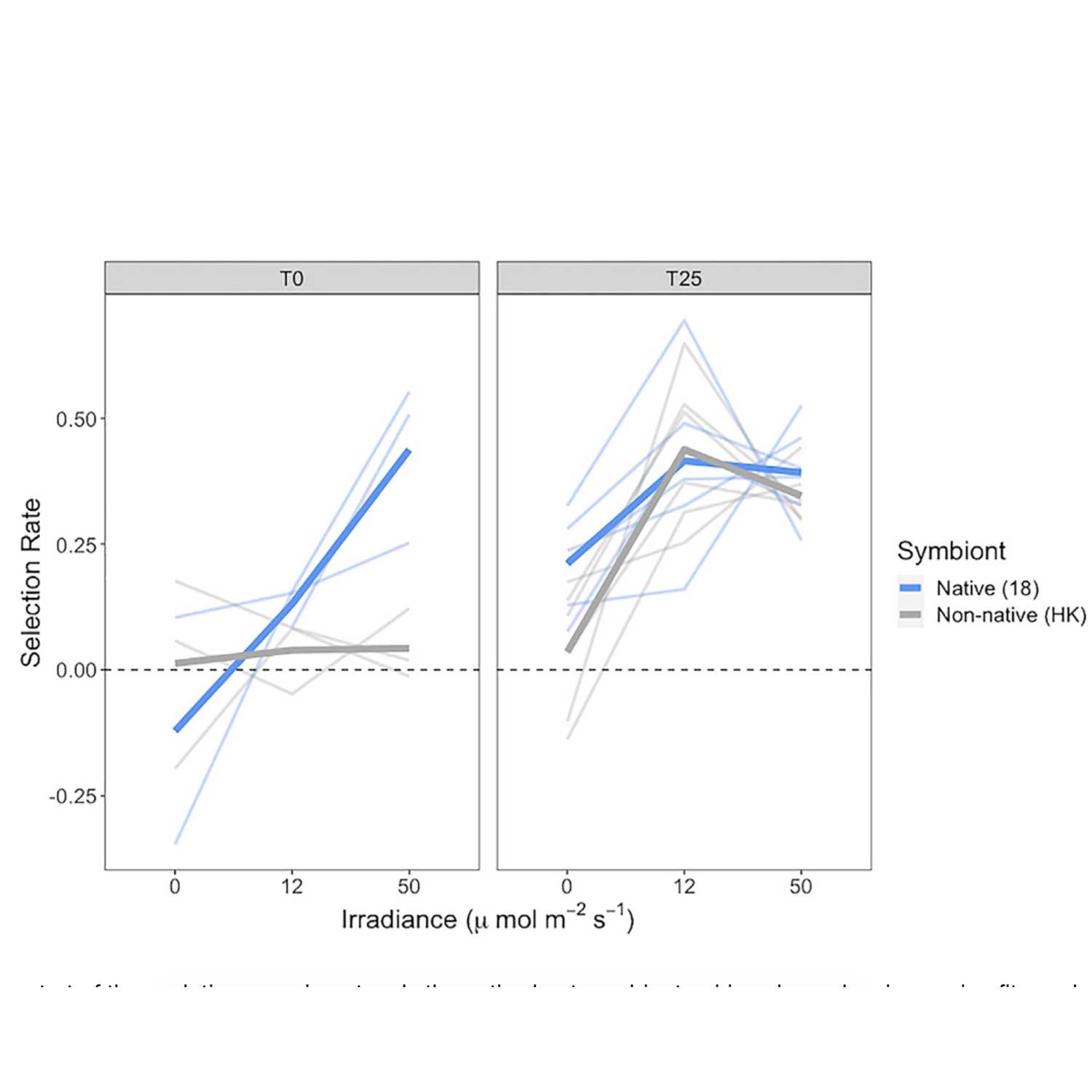
Papers Aloud#29 - Rapid compensatory evolution - Sørensen et al., 2021 - Current BiologyTitle: Rapid compensatory evolution can rescue low fitness symbioses following partner switching. Authors: Megan E.S. Sørensen, A. Jamie Wood, Duncan D. Cameron, Michael A. Brockhurst. Citation: Sørensen,M.E.S. et al. (2021) Rapid compensatory evolution can rescue low fitness symbioses following partner switching. Curr. Biol.. Link to article
2021-07-3111 min
Papers Aloud#28 - At the Gate of Mutualism - Renoz et al., 2021 - Frontiers in MicrobiologyTitle: At the Gate of Mutualism: Identification of Genomic Traits Predisposing to Insect-Bacterial Symbiosis in Pathogenic Strains of the Aphid Symbiont Serratia symbiotica. Authors: François Renoz, Vincent Foray, Jérôme Ambroise, Patrice Baa-Puyoulet, Bertrand Bearzatto, Gipsi Lima Mendez, Alina S. Grigorescu, Jacques Mahillon, Patrick Mardulyn, Jean-Luc Gala, Federica Calevro, and Thierry Hance. Citation: Renoz, F. et al. (2021) At the Gate of Mutualism: Identification of Genomic Traits Predisposing to Insect-Bacterial Symbiosis in Pathogenic Strains of the Aphid Symbiont Serratia symbiotica. Front. Cell. Infect. Microbiol., 11, 660007. Link to article
2021-07-2448 min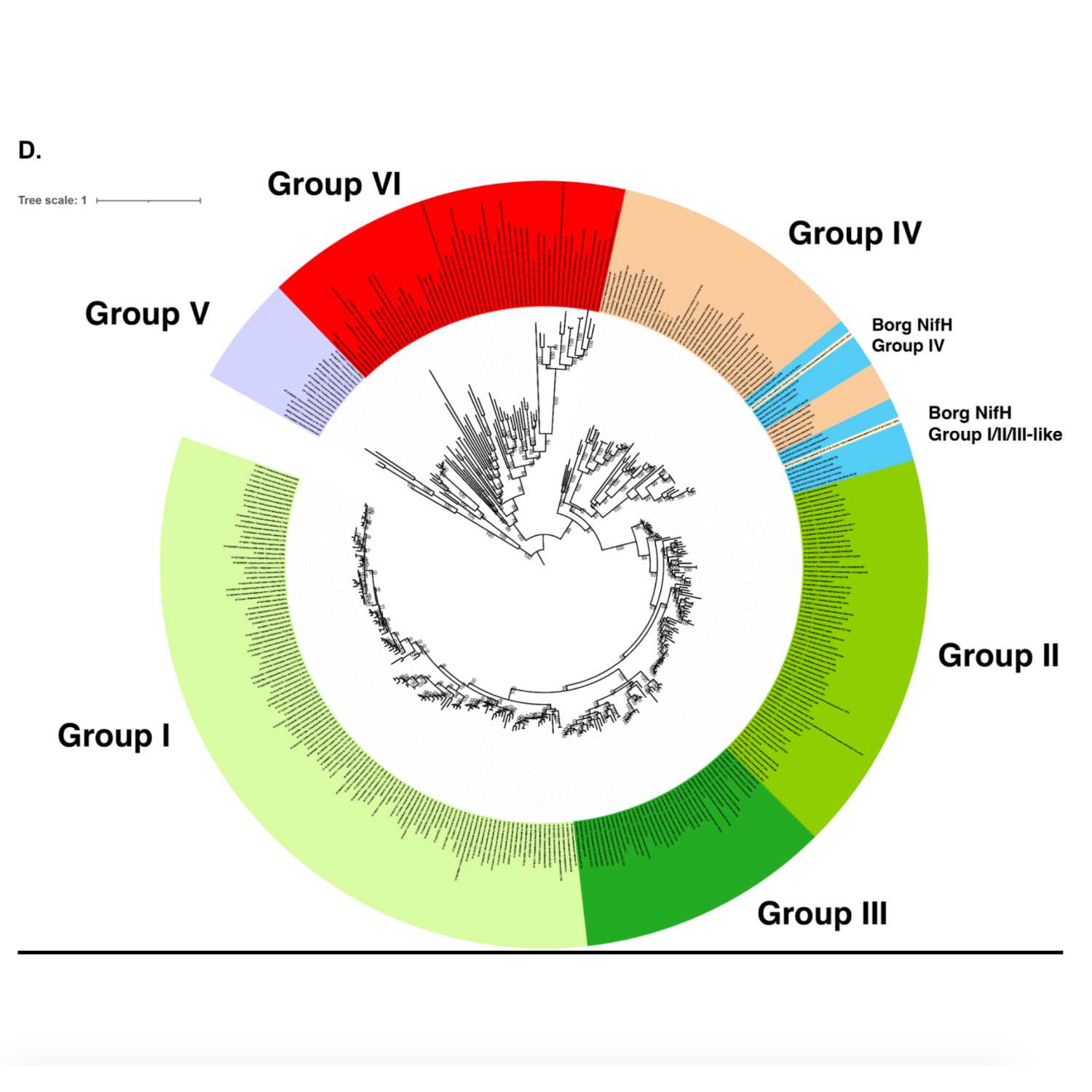
Papers Aloud#27 - Borgs - Al-Shayeb et al., 2021 - BioRxiv preprintTitle: Borgs are giant extrachromosomal elements with the potential to augment methane oxidation. Authors: Basem Al-Shayeb, Marie C. Schoelmerich, Jacob West-Roberts, Luis E. Valentin-Alvarado, Rohan Sachdeva, Susan Mullen, Alexander Crits-Christoph, Michael J. Wilkins, Kenneth H. Williams, Jennifer A. Doudna, Jillian F. Banfield. Citation: Al-Shayeb, B. et al. (2021) Borgs are giant extrachromosomal elements with the potential to augment methane oxidation. bioRxiv, 2021.07.10.451761. Link to article
2021-07-2207 min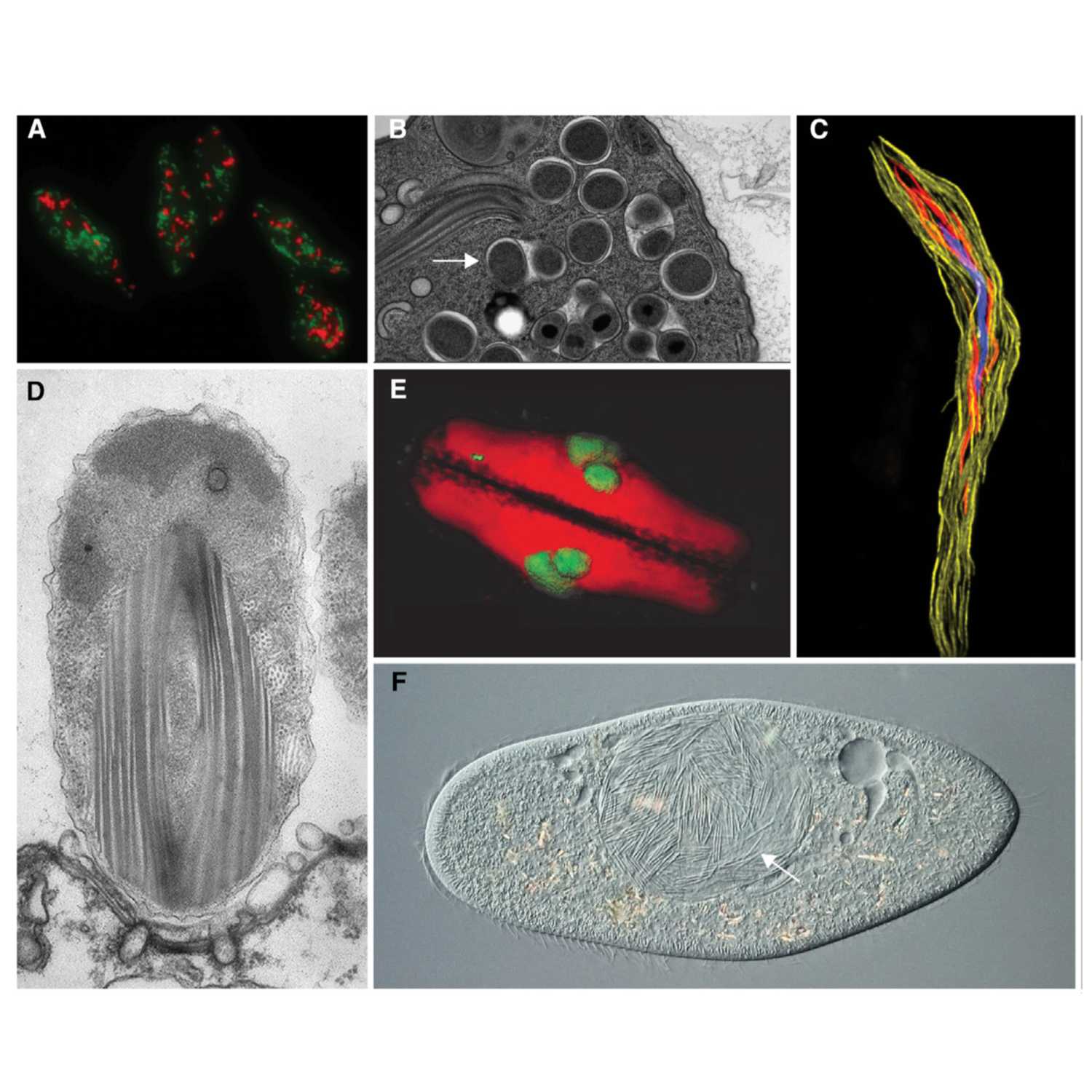
Papers Aloud#26 - Bacterial and archaeal symbioses with protists - Husnik et al., 2021 - Current BiologyTitle: Bacterial and archaeal symbioses with protists. Authors: Filip Husnik, Daria Tashyreva, Vittorio Boscaro, Emma E. George, Julius Lukes, and Patrick J. Keeling. Citation: Husnik,F. et al. (2021) Bacterial and archaeal symbioses with protists. Curr. Biol., 31, R862–R877. Link to article
2021-07-2152 min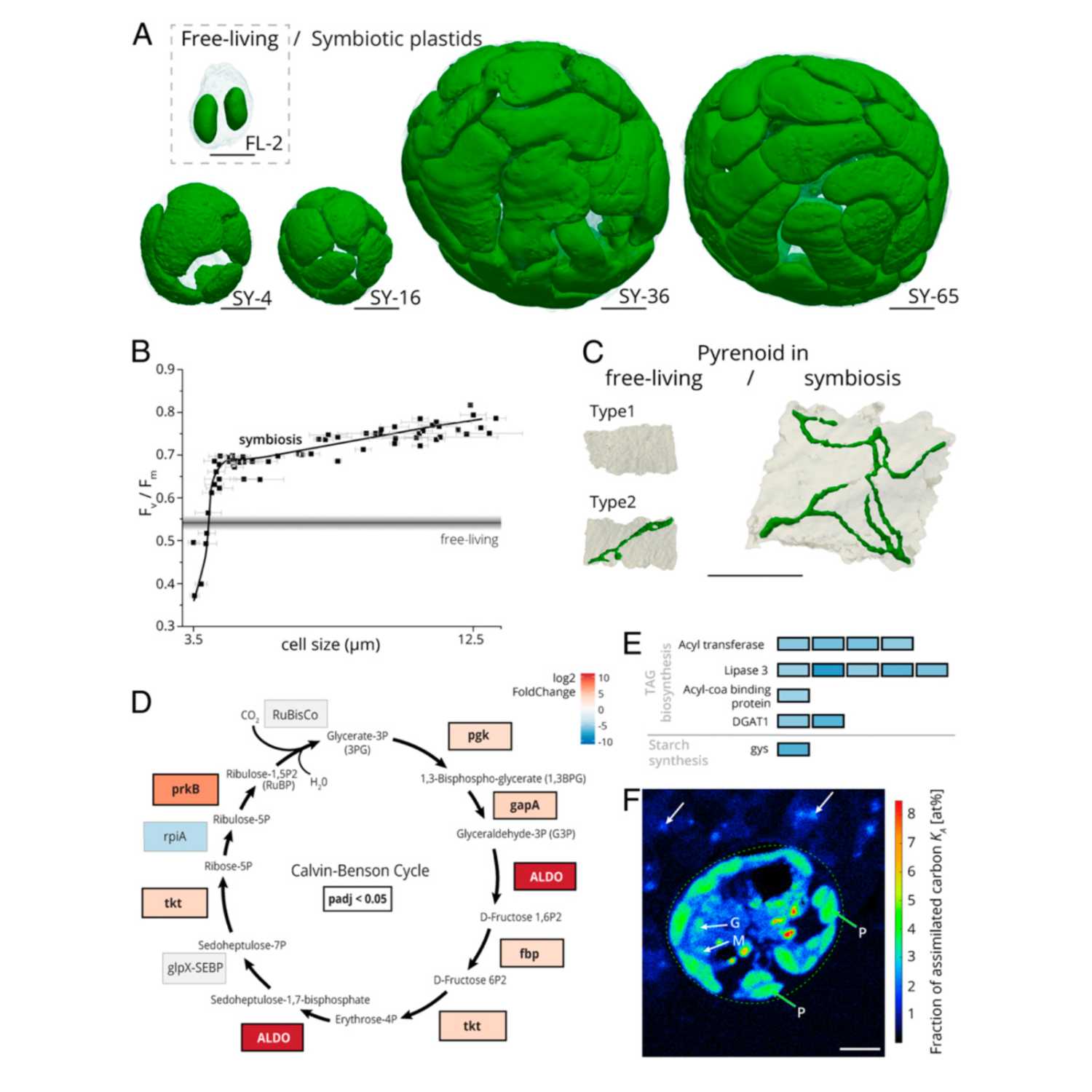
Papers Aloud#25 - Cytoklepty in the plankton - Uwizeye et al., 2021 - PNASTitle: Cytoklepty in the plankton: A host strategy to optimize the bioenergetic machinery of endosymbiotic algae. Authors: Clarisse Uwizeye, Margaret Mars Brisbin, Benoit Gallet, Fabien Chevalier, Charlotte LeKieffre, Nicole L. Schieber, Denis Falconet, Daniel Wangpraseurt, Lukas Schertel, Hryhoriy Stryhanyuk, Niculina Musat, Satoshi Mitarai, Yannick Schwab, Giovanni Finazzi, and Johan Decelle. Citation: Uwizeye,C. et al. (2021) Cytoklepty in the plankton: A host strategy to optimize the bioenergetic machinery of endosymbiotic algae. Proc. Natl. Acad. Sci. U. S. A., 118. Link to article
2021-07-1415 min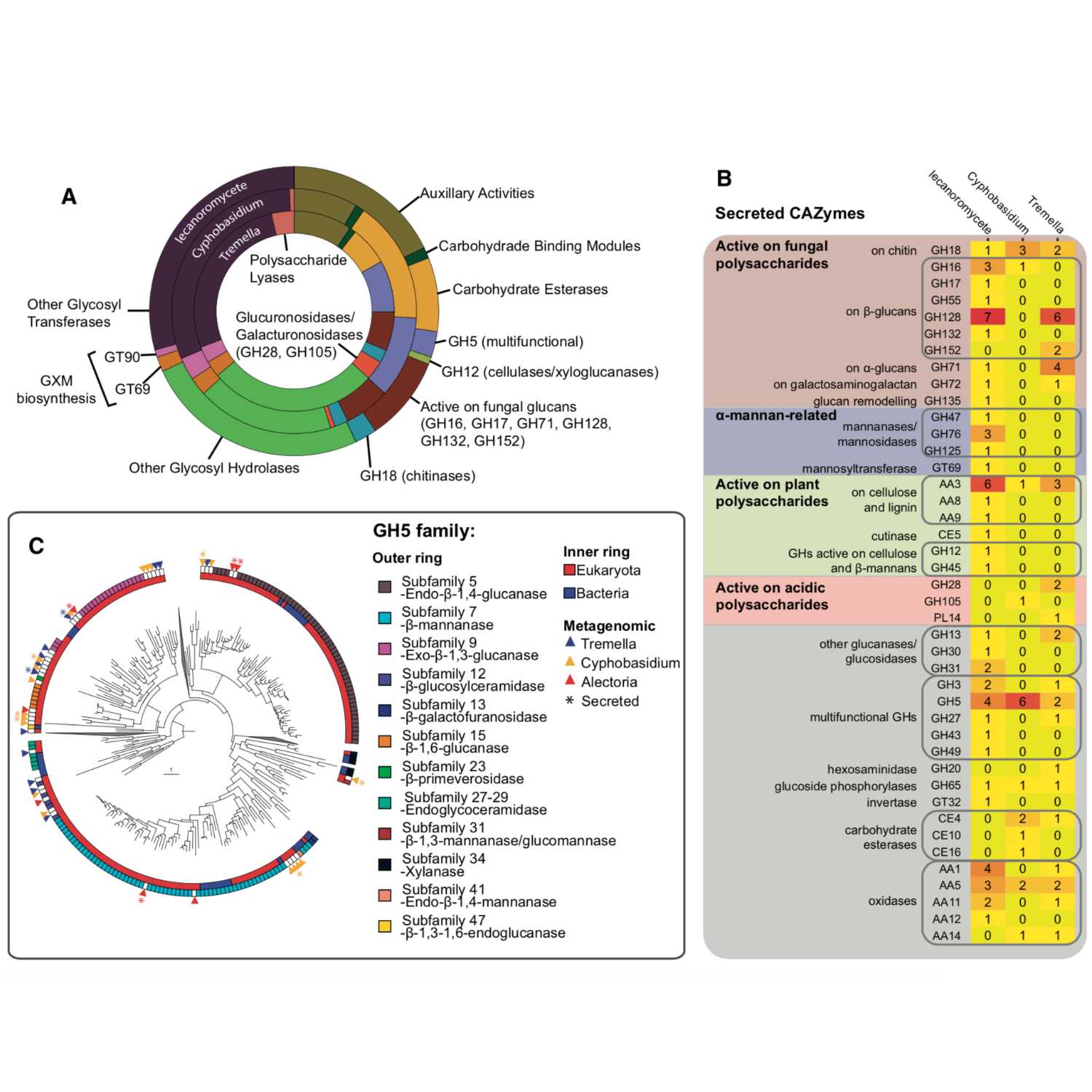
Papers Aloud#24 - Fungal Symbionts in a Lichen Symbiosis - Tagirdzhanova et al., 2021 - GBETitle: Predicted Input of Uncultured Fungal Symbionts to a Lichen Symbiosis from Metagenome-Assembled Genomes. Authors: Gulnara Tagirdzhanova, Paul Saary, Jeffrey P. Tingley, David Dıaz-Escando, D. Wade Abbott, Robert D. Finn2 and Toby Spribille. Citation: Tagirdzhanova, G. et al. (2021) Predicted Input of Uncultured Fungal Symbionts to a Lichen Symbiosis from Metagenome-Assembled Genomes. Genome Biol. Evol., 13. Link to article
2021-07-1328 min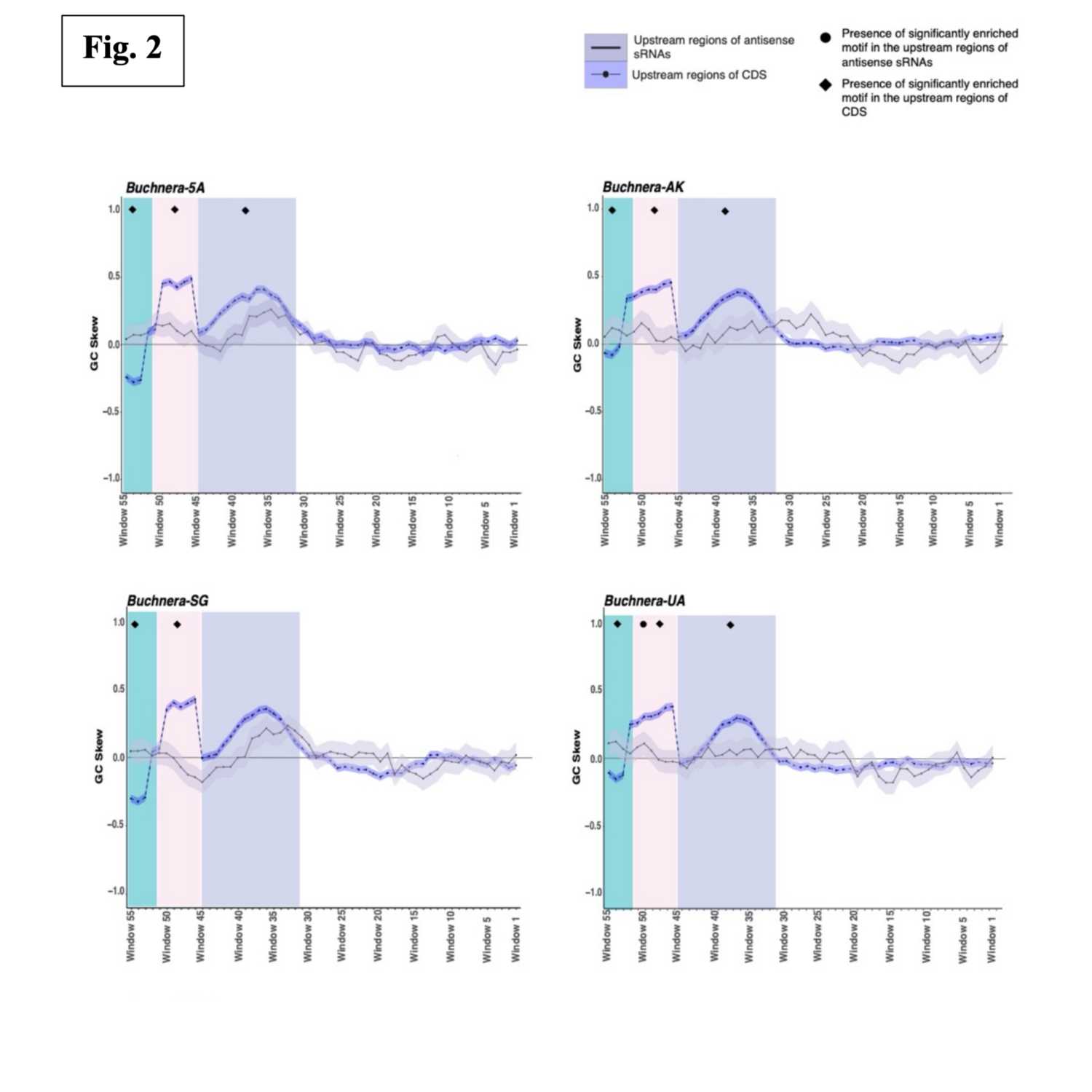
Papers Aloud#23 - Small RNAs in divergent host-restricted bacteria - Thairu et al., 2021 - MBETitle: Natural selection shapes maintenance of orthologous sRNAs in divergent host-restricted bacterial genomes. Authors: Margaret W. Thairu, Venkata Rama Sravani Meduri, Patrick H. Degnan, Allison K. Hansen. Citation: Thairu, M.W. et al. (2021) Natural selection shapes maintenance of orthologous sRNAs in divergent host-restricted bacterial genomes. Mol. Biol. Evol.Link to article
2021-07-0925 min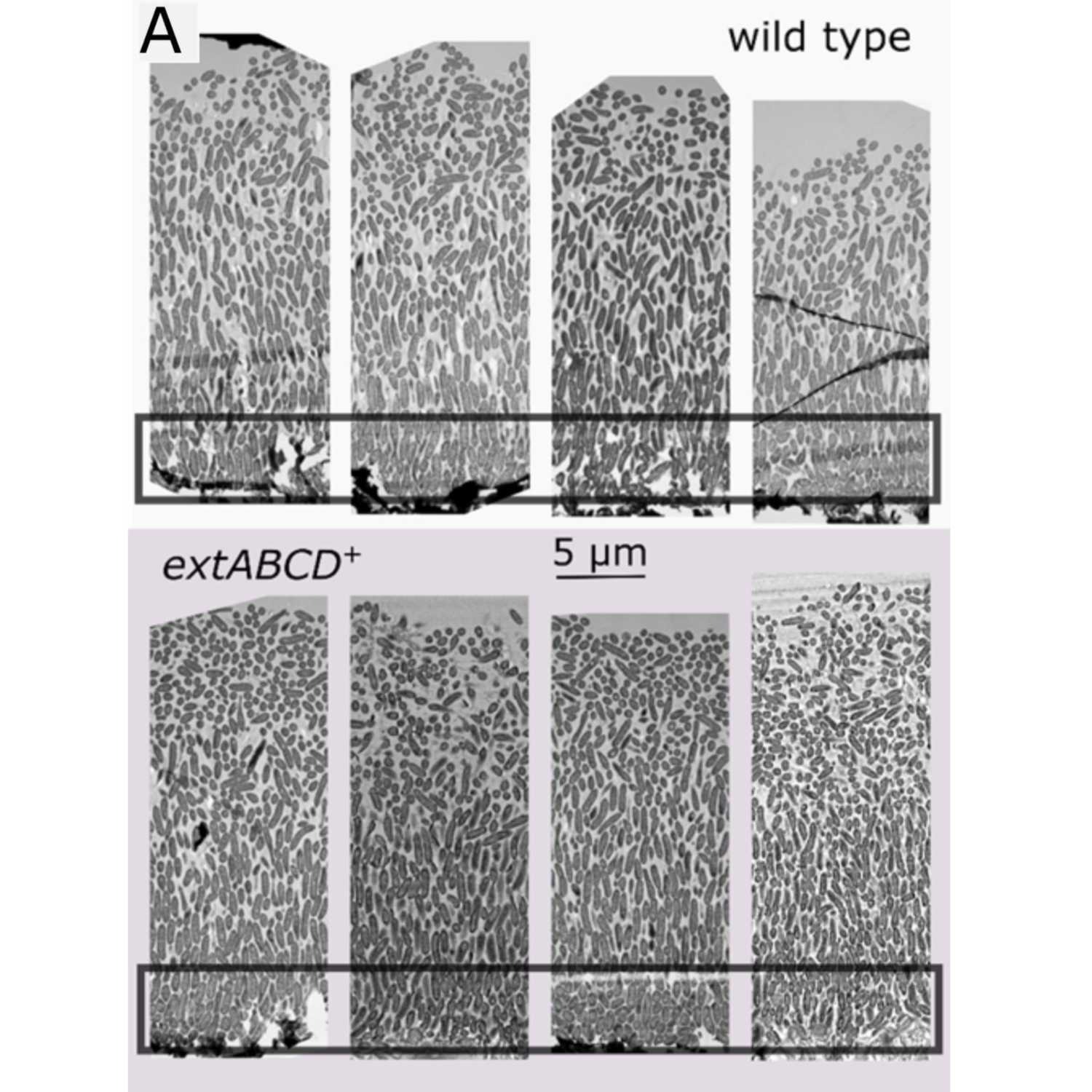
Papers Aloud#22 - Streamlined extracellular electron transfer pathway - Otero et al., 2021 - AEMTitle: Evidence of streamlined extracellular electron transfer pathway from biofilm structure, metabolic stratification, and long-range electron transfer parameters.Authors: Fernanda Jiménez Otero, Grayson L. Chadwick, Matthew D. Yates, Rebecca L. Mickol, Scott H. Saunders, Sarah M. Glaven, Jeffrey A. Gralnick, Dianne K. Newman, Leonard M. Tender, Victoria J. Orphan, Daniel R. Bond.Citation: Jiménez Otero, F. et al. (2021) Evidence of streamlined extracellular electron transfer pathway from biofilm structure, metabolic stratification, and long-range electron transfer parameters. Appl. Environ. Microbiol., AEM0070621.Link to article
2021-07-0722 min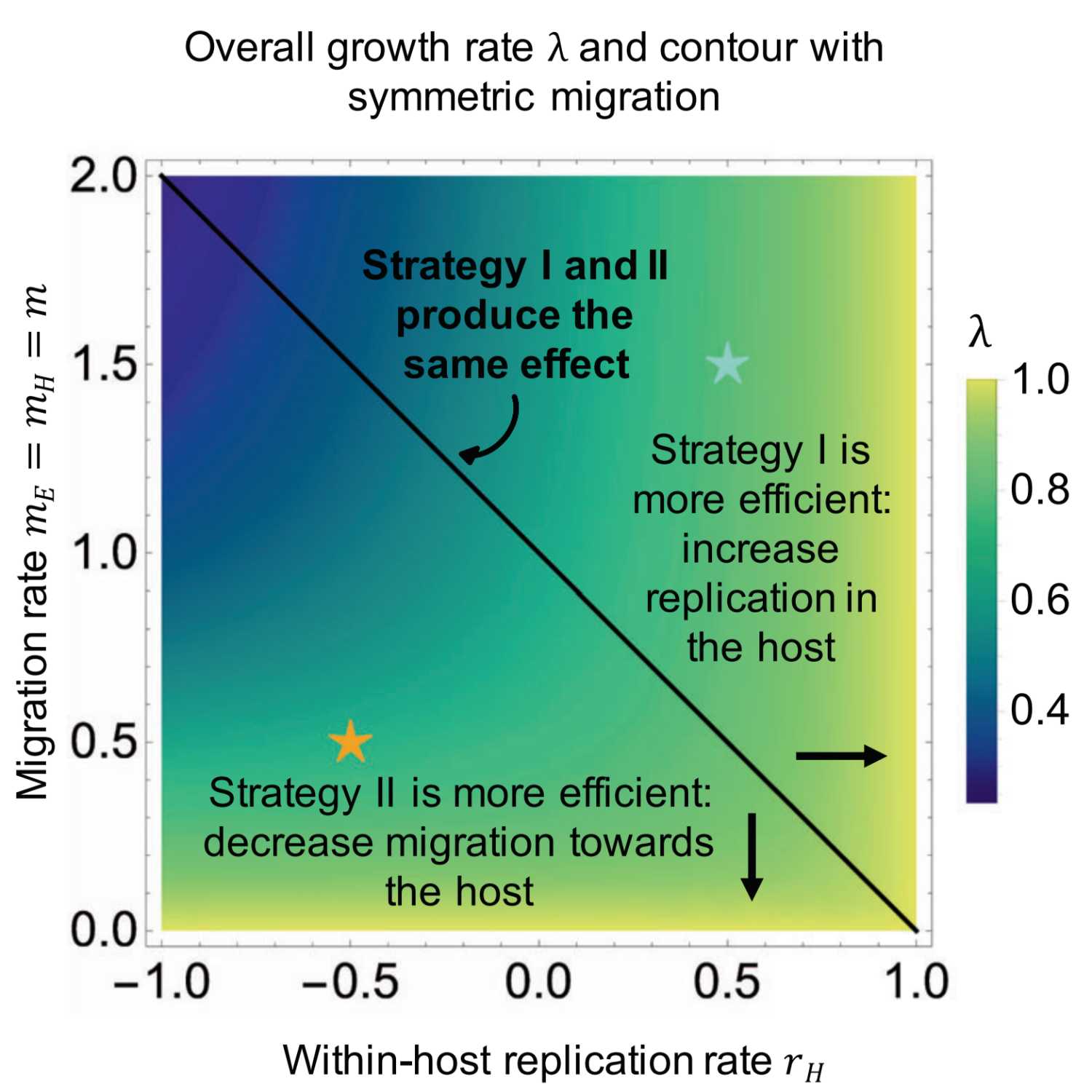
Papers Aloud#21 - Modeling host-associating microbes under selection - Bansept et al., 2021 - ISMETitle: Modeling host-associating microbes under selectionAuthors: Florence Bansept, Nancy Obeng, Hinrich Schulenburg, and Arne TraulsenCitation: Bansept, F. et al. (2021) Modeling host-associating microbes under selection. ISME J.Link to article
2021-06-2521 min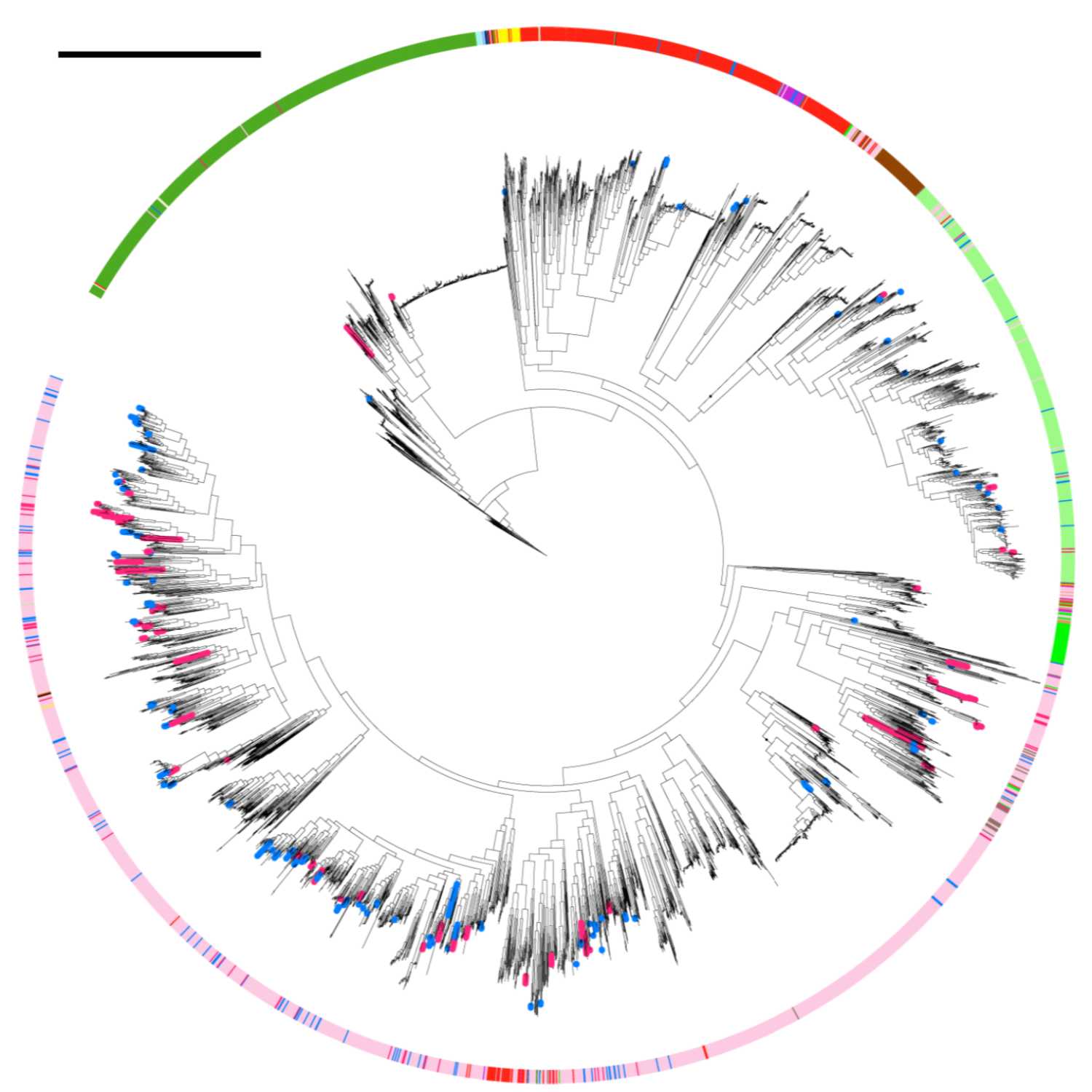
Papers Aloud#20 - Ancient microbial genomes from the human gut - Wibowo et al., 2021 - NatureTitle: Reconstruction of ancient microbial genomes from the human gutAuthors: Marsha C. Wibowo, Zhen Yang, Maxime Borry, Alexander Hübner, Kun D. Huang, Braden T. Tierney, Samuel Zimmerman, Francisco Barajas-Olmos, Cecilia Contreras-Cubas, Humberto García-Ortiz, Angélica Martínez-Hernández, Jacob M. Luber, Philipp Kirstahler, Tre Blohm, Francis E. Smiley, Richard Arnold, Sonia A. Ballal, Sünje Johanna Pamp, Julia Russ, Frank Maixner, Omar Rota-Stabelli, Nicola Segata, Karl Reinhard, Lorena Orozco, Christina Warinner, Meradeth Snow, Steven LeBlanc, and Aleksandar D. KosticCitation: Wibowo, M.C. et al. (2021) Reconstruction of ancient microbial genomes from the human...
2021-06-2010 min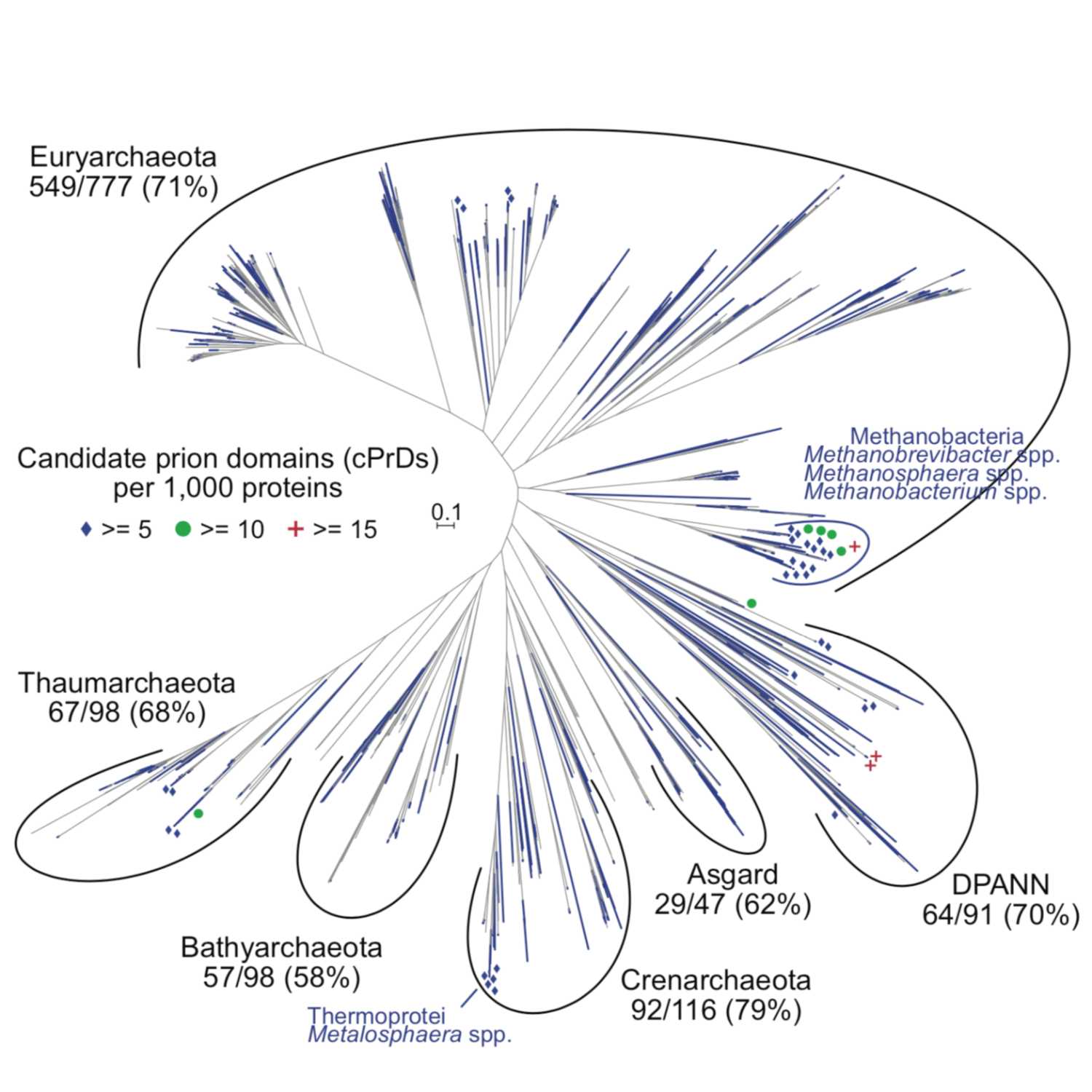
Papers Aloud#19 - The Hunt for Ancient Prions - Zajkowski et al., 2021 - MBETitle: The Hunt for Ancient Prions: Archaeal Prion-Like Domains Form Amyloid-Based Epigenetic ElementsAuthors: Tomasz Zajkowski, Michael D. Lee, Shamba S. Mondal, Amanda Carbajal, Robert Dec, Patrick D. Brennock, Radoslaw W. Piast, Jessica E. Snyder, Nicholas B. Bense, Wojciech Dzwolak, Daniel F. Jarosz, and Lynn J. RothschildCitation: Zajkowski, T. et al. (2021) The Hunt for Ancient Prions: Archaeal Prion-Like Domains Form Amyloid-Based Epigenetic Elements. Mol. Biol. Evol., 38, 2088–2103.Link to article
2021-06-2026 min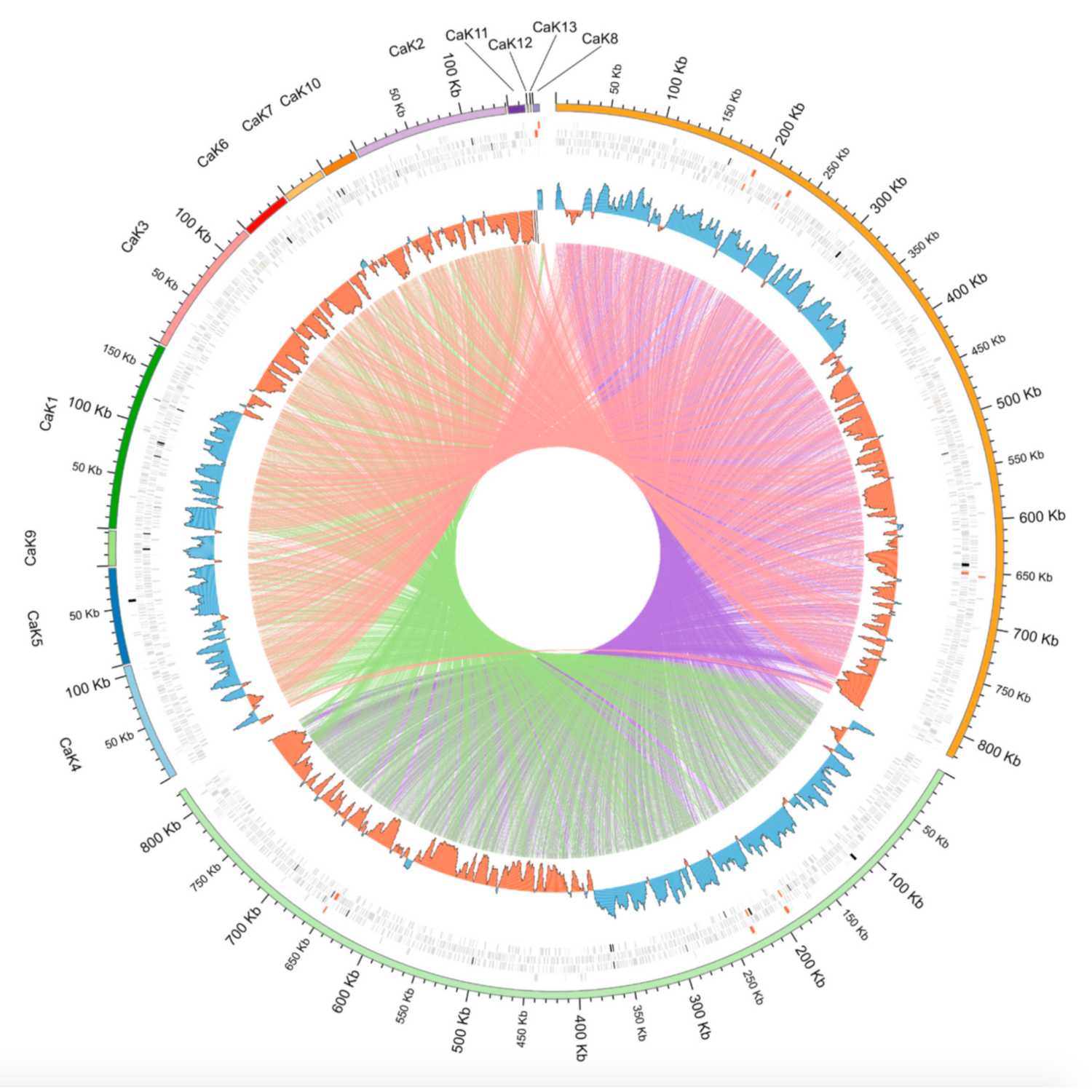
Papers Aloud#18 - Endosymbiont Capture - Skalický et al., 2021 - PathogensTitle: Endosymbiont Capture, a Repeated Process of Endosymbiont Transfer with Replacement in Trypanosomatids Angomonas spp.Authors: Tomáš Skalický, João M. P. Alves , Anderson C. Morais, Jana Režnarová, Anzhelika Butenko, Julius Lukeš, Myrna G. Serrano, Gregory A. Buck, Marta M. G. Teixeira, Erney P. Camargo, Mandy Sanders, James A. Cotton, Vyacheslav Yurchenko, and Alexei Y. KostygovCitation: Skalický,T. et al. (2021) Endosymbiont Capture, a Repeated Process of Endosymbiont Transfer with Replacement in Trypanosomatids Angomonas spp. Pathogens, 10, 702.Link to article
2021-06-1916 min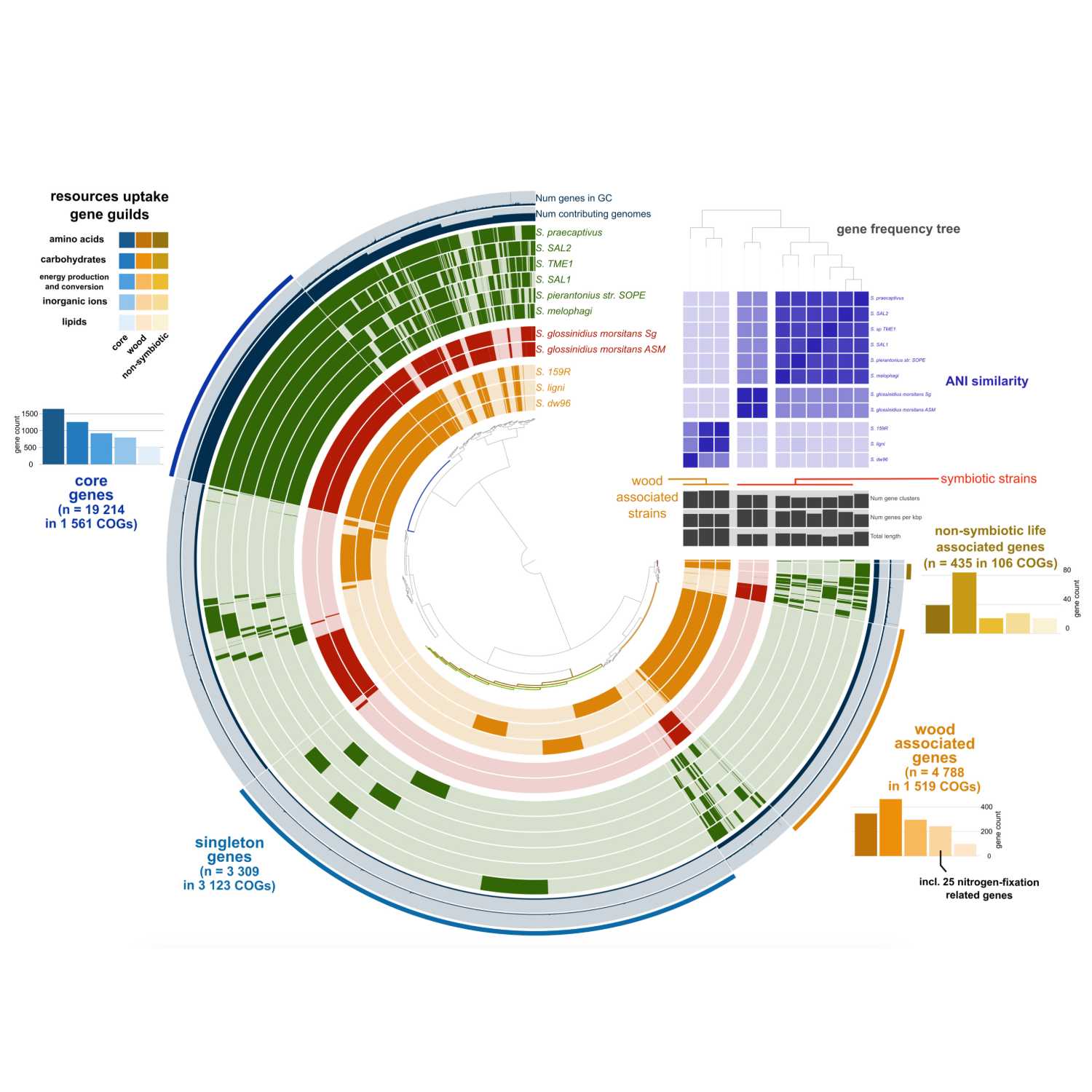
Papers Aloud#17 - Sodalis: From Insect Symbionts to Inhabitants of Decomposing Deadwood - Tláskal et al., 2021 - Frontiers in MicrobiologyTitle: Ecological Divergence Within the Enterobacterial Genus Sodalis: From Insect Symbionts to Inhabitants of Decomposing DeadwoodAuthors: Vojtēch Tláskal, Victor Satler Pylro, Lucia Žifčáková, and Petr BaldrianCitation: Tláskal,V. et al. (2021) Ecological Divergence Within the Enterobacterial Genus Sodalis: From Insect Symbionts to Inhabitants of Decomposing Deadwood. Front. Microbiol., 12, 1499.Link to article
2021-06-1919 min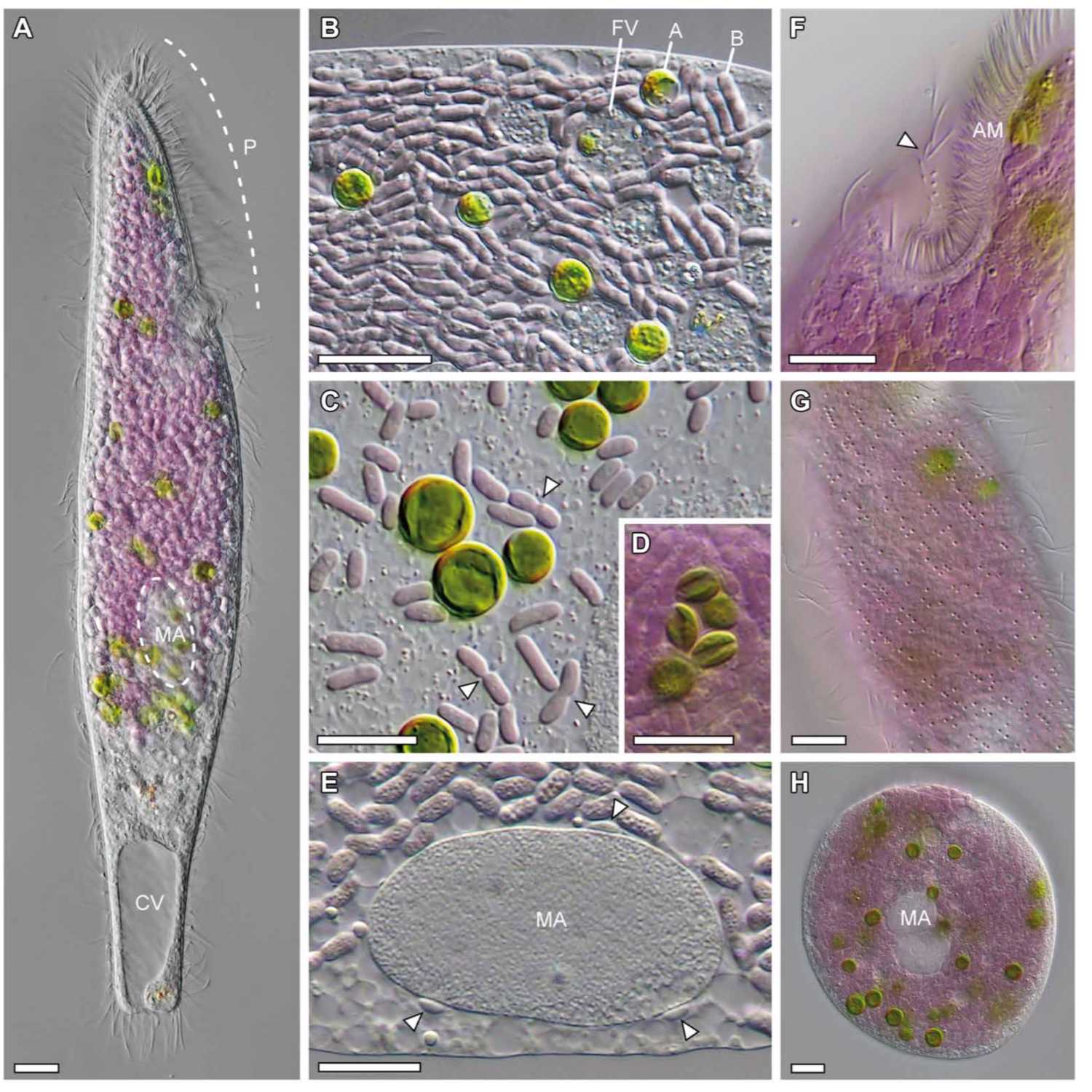
Papers Aloud#16 - A microbial eukaryote with purple bacteria and green algae endosymbionts - Muñoz-Gómez et al., 2021 - Science AdvancesTitle: A microbial eukaryote with a unique combination of purple bacteria and green algae as endosymbiontsAuthors: Sergio A. Muñoz-Gómez, Martin Kreutz, Sebastian HessCitation: Muñoz-Gómez, S.A. et al. (2021) A microbial eukaryote with a unique combination of purple bacteria and green algae as endosymbionts. Sci Adv, 7.Link to article
2021-06-1528 min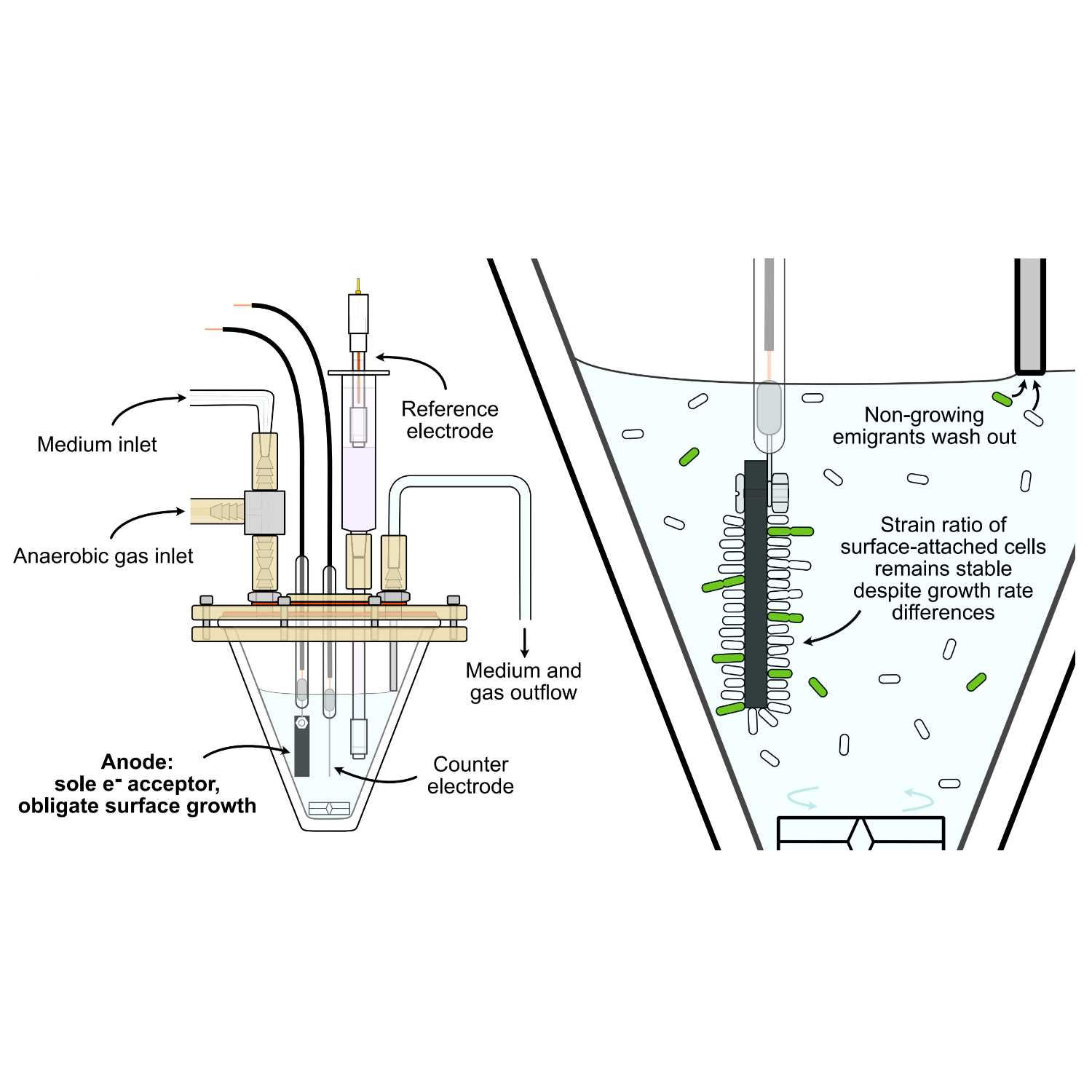
Papers Aloud#15 - Shewanella electrode biofilms - Kees et al., 2021 - Nature CommunicationsTitle: Survival of the first rather than the fittest in a Shewanella electrode biofilmAuthors: Eric D. Kees, Caleb E. Levar, Stephen P. Miller, Daniel R. Bond, Jeffrey A. Gralnick, and Antony M. DeanCitation: Kees, E.D. et al. (2021) Survival of the first rather than the fittest in a Shewanella electrode biofilm. Commun Biol, 4, 536.Link to article
2021-06-0317 min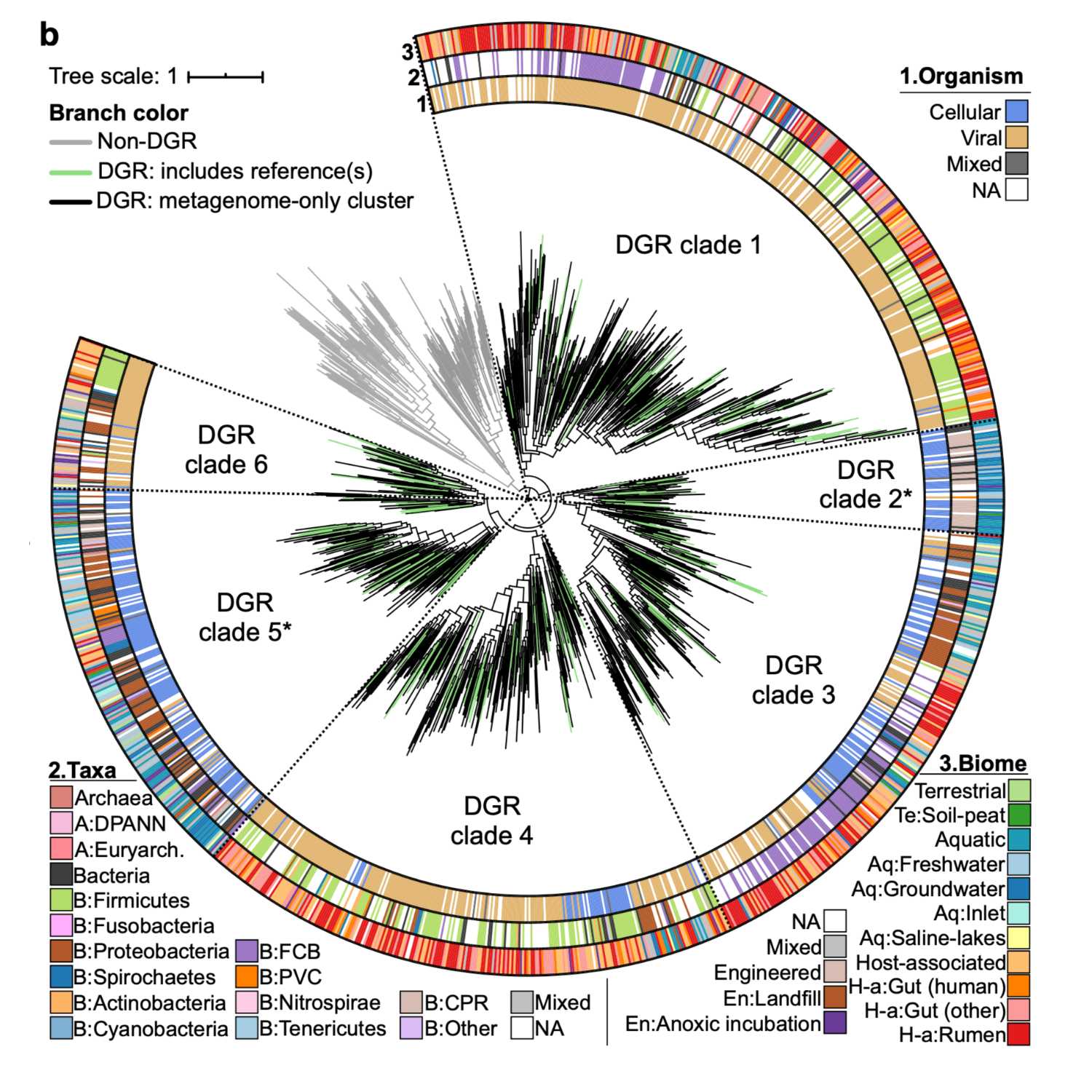
Papers Aloud#14 - Hypermutation in the global microbiome - Roux et al., 2021 - Nature CommunicationsTitle: Ecology and molecular targets of hypermutation in the global microbiomeAuthors: Simon Roux , Blair G. Paul, Sarah C. Bagby, Stephen Nayfach, Michelle A. Allen, Graeme Attwood, Ricardo Cavicchioli, Ludmila Chistoserdova, Robert J. Gruninger, Steven J. Hallam, Maria E. Hernandez, Matthias Hess, Wen-Tso Liu, Tim A. McAllister, Michelle A. O’Malley, Xuefeng Peng, Virginia I. Rich, Scott R. Saleska, and Emiley A. Eloe-FadroshCitation: Roux, S. et al. (2021) Ecology and molecular targets of hypermutation in the global microbiome. Nat. Commun., 12, 3076.Link to article
2021-06-0310 min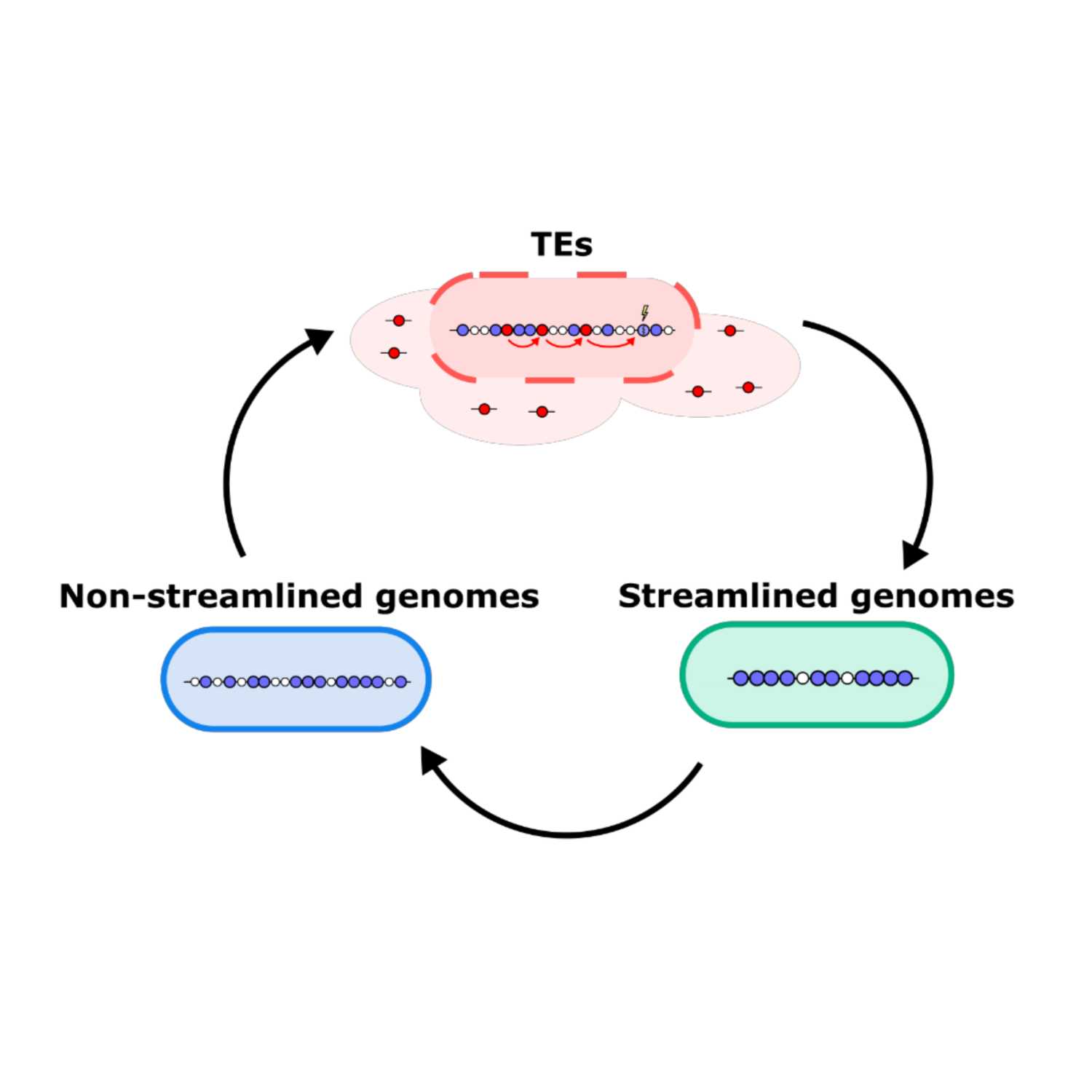
Papers Aloud#13 - Transposable elements and genome streamlining - Dijk et al., 2021 - bioRxiv preprintTitle: Transposable elements drive the evolution of genome streamliningAuthors: Bram van Dijk, Frederic Bertels, Lianne Stolk, Nobuto Takeuchi, and Paul B. RaineyCitation: van Dijk, B. et al. (2021) Transposable elements drive the evolution of genome streamlining. bioRxiv, 2021.05.29.446280.Link to article
2021-06-0314 min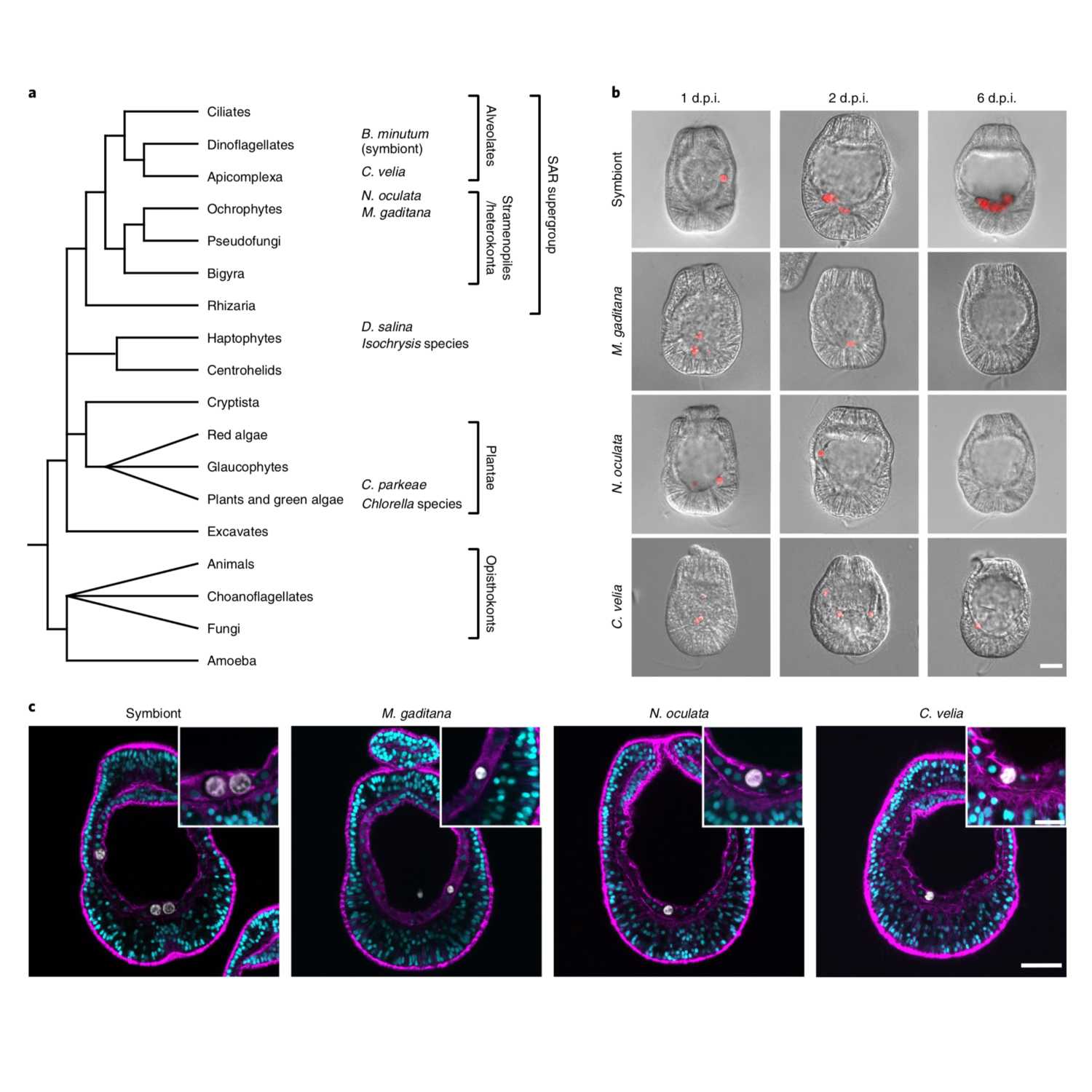
Papers Aloud#12 - Dinoflagellate symbionts - Jacobovitz et al., 2021 - Nature MicrobiologyTitle: Dinoflagellate symbionts escape vomocytosis by host cell immune suppressionAuthors: Marie R. Jacobovitz, Sebastian Rupp, Philipp A. Voss, Ira Maegele, Sebastian G. Gornik, and Annika GuseCitation: Jacobovitz, M.R. et al. (2021) Dinoflagellate symbionts escape vomocytosis by host cell immune suppression. Nature Microbiology, 6, 769–782.Link to article
2021-05-2929 min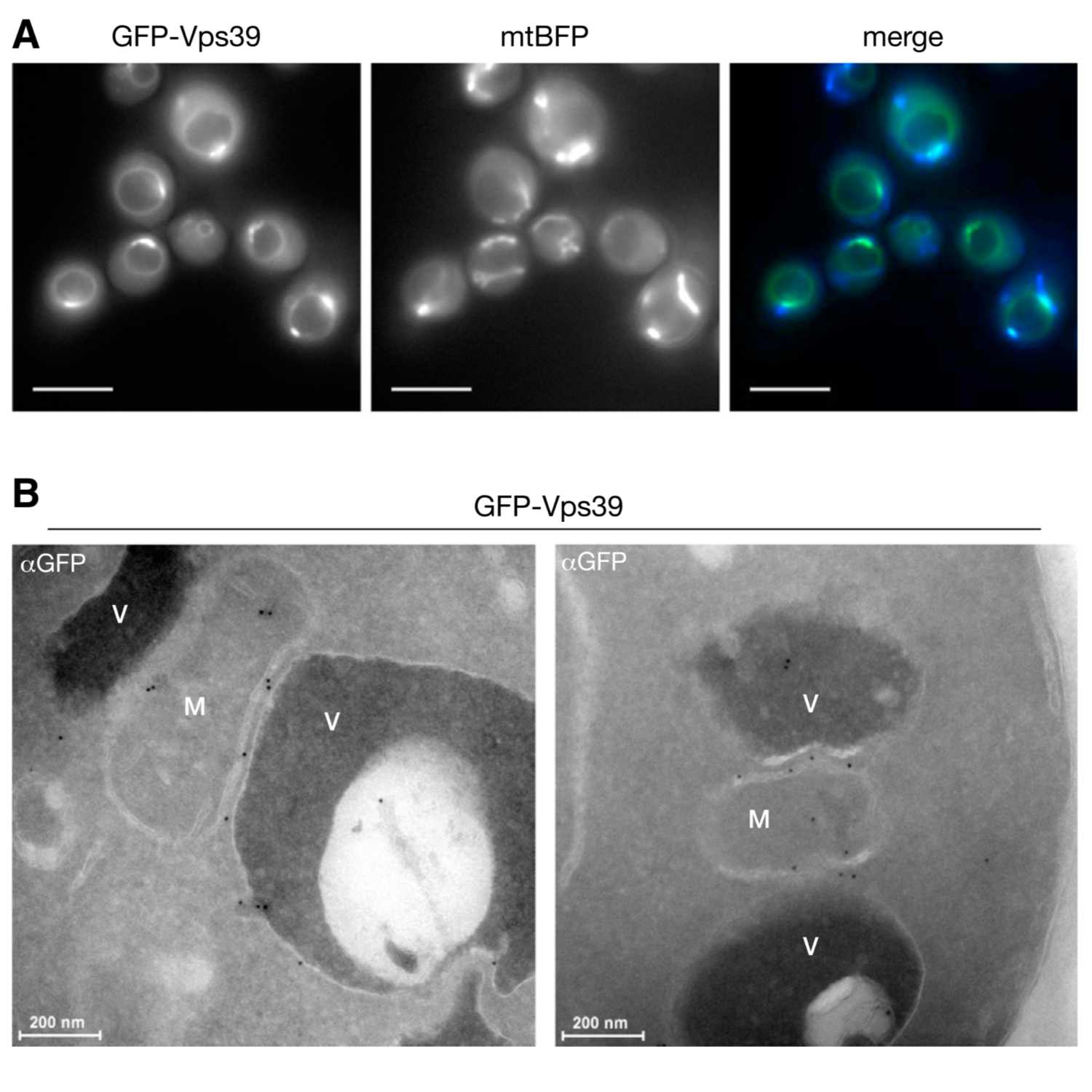
Papers Aloud#11 - Interface between Vacuoles and Mitochondria - Elbaz-Alon et al., 2014 - Cell PressTitle: A Dynamic Interface between Vacuoles and Mitochondria in Yeast. Authors: Yael Elbaz-Alon, Eden Rosenfeld-Gur, Vera Shinder, Anthony H. Futerman, Tamar Geiger, and Maya Schuldiner. Citation: Elbaz-Alon, Y. et al. (2014) A dynamic interface between vacuoles and mitochondria in yeast. Dev. Cell, 30, 95–102. Link to article
2021-05-2811 min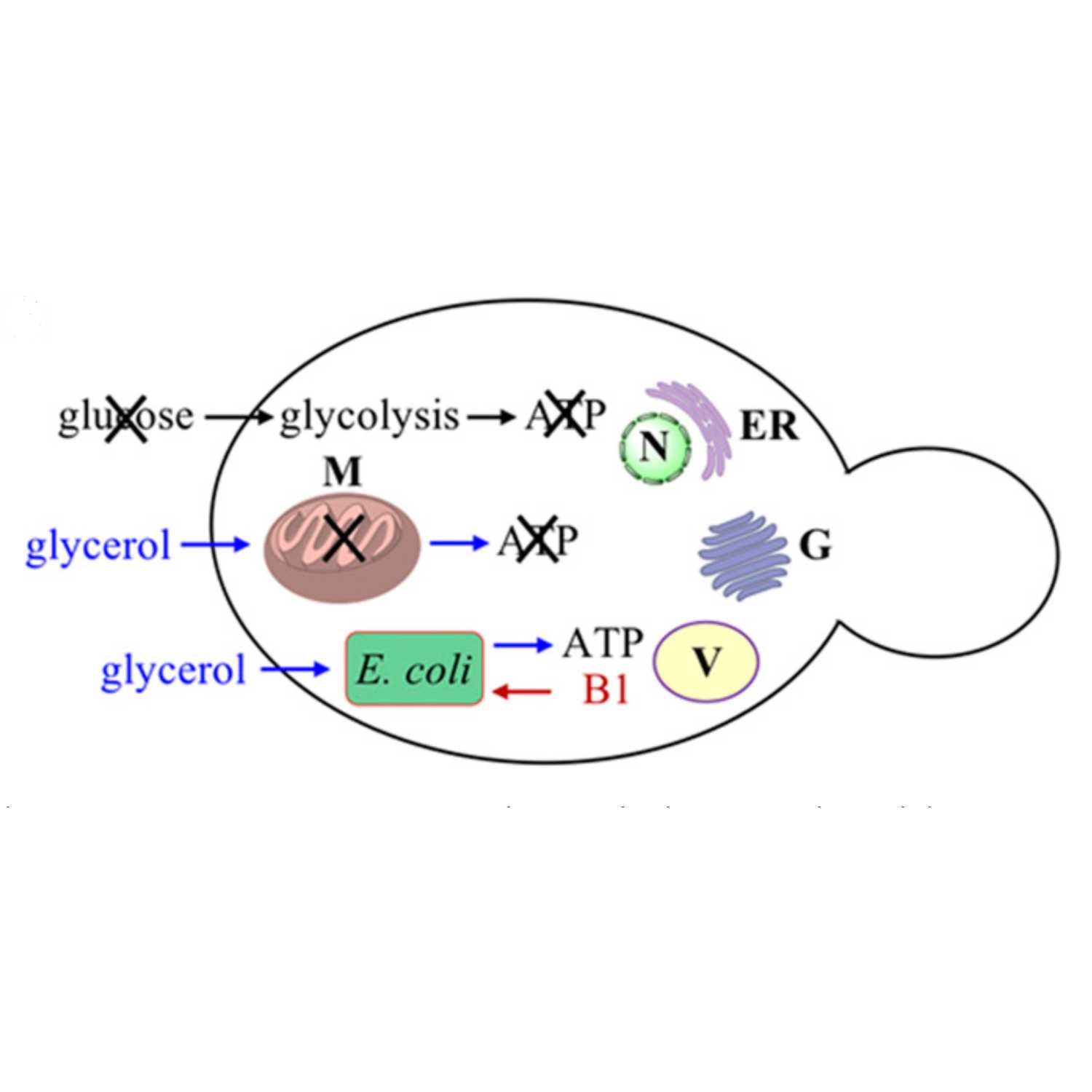
Papers Aloud#10 - Engineering yeast endosymbionts - Mehta et al., 2018 - PNASTitle: Engineering yeast endosymbionts as a step toward the evolution of mitochondria Authors: Angad P. Mehta, Lubica Supekova, Jian-Hua Chen, Kersi Pestonjamasp, Paul Webster, Yeonjin Ko, Scott C. Henderson, Gerry McDermott, Frantisek Supeke, and Peter G. SchultzCitation: Mehta,A.P. et al. (2018) Engineering yeast endosymbionts as a step toward the evolution of mitochondria. Proc. Natl. Acad. Sci. U. S. A., 115, 11796–11801.Link to article
2021-05-1510 min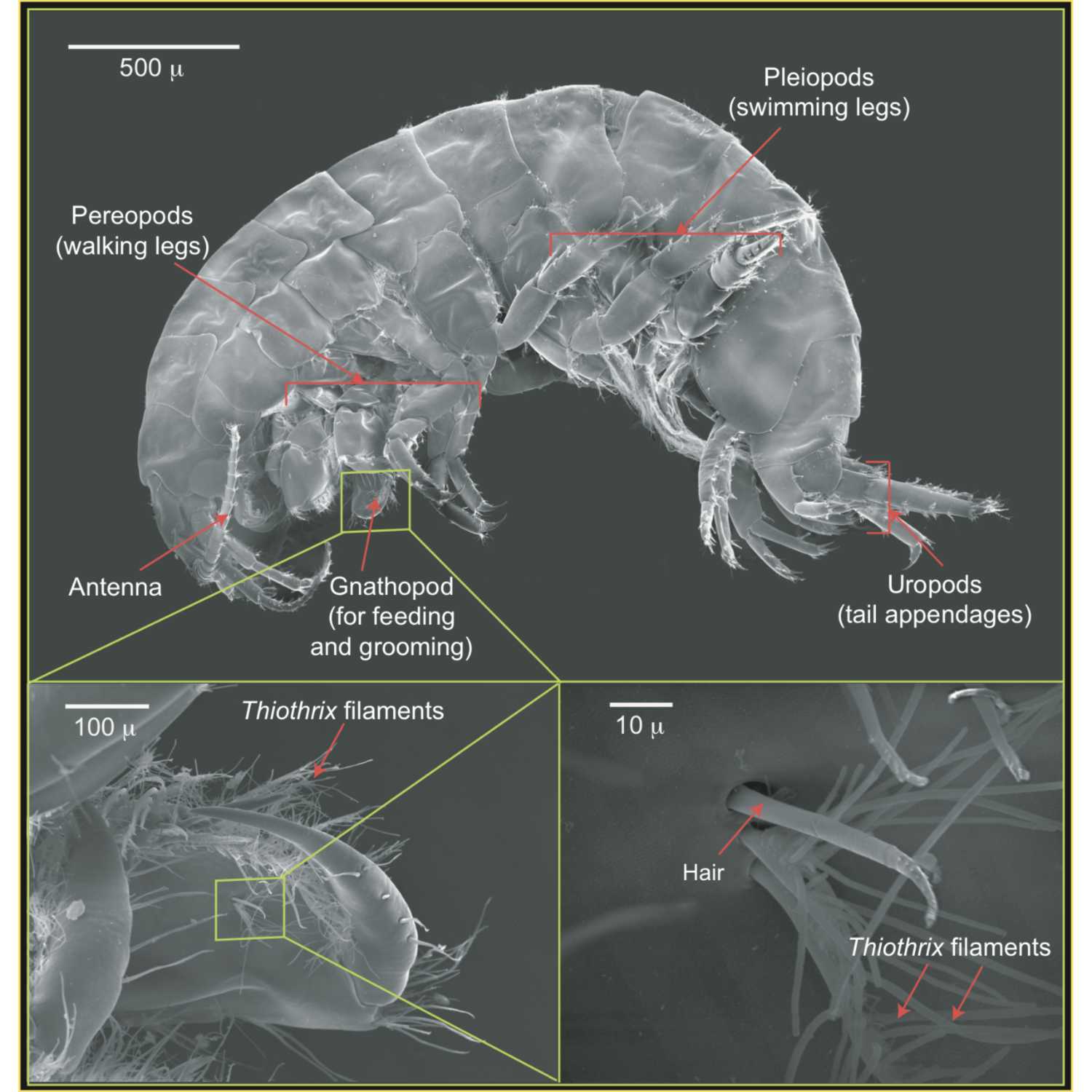
Papers Aloud#9 - Novel Chemoautotrophic Symbiosis - Dattagupta et al., 2009 - ISMETitle: A novel symbiosis between chemoautotrophic bacteria and a freshwater cave amphipodAuthors: Sharmishtha Dattagupta, Irene Schaperdoth, Alessandro Montanari, Sandro Mariani, Noriko Kita, John W Valley, and Jennifer L MacaladyCitation: Dattagupta, S. et al. (2009) A novel symbiosis between chemoautotrophic bacteria and a freshwater cave amphipod. ISME J., 3, 935–943.Link to article
2021-05-1421 min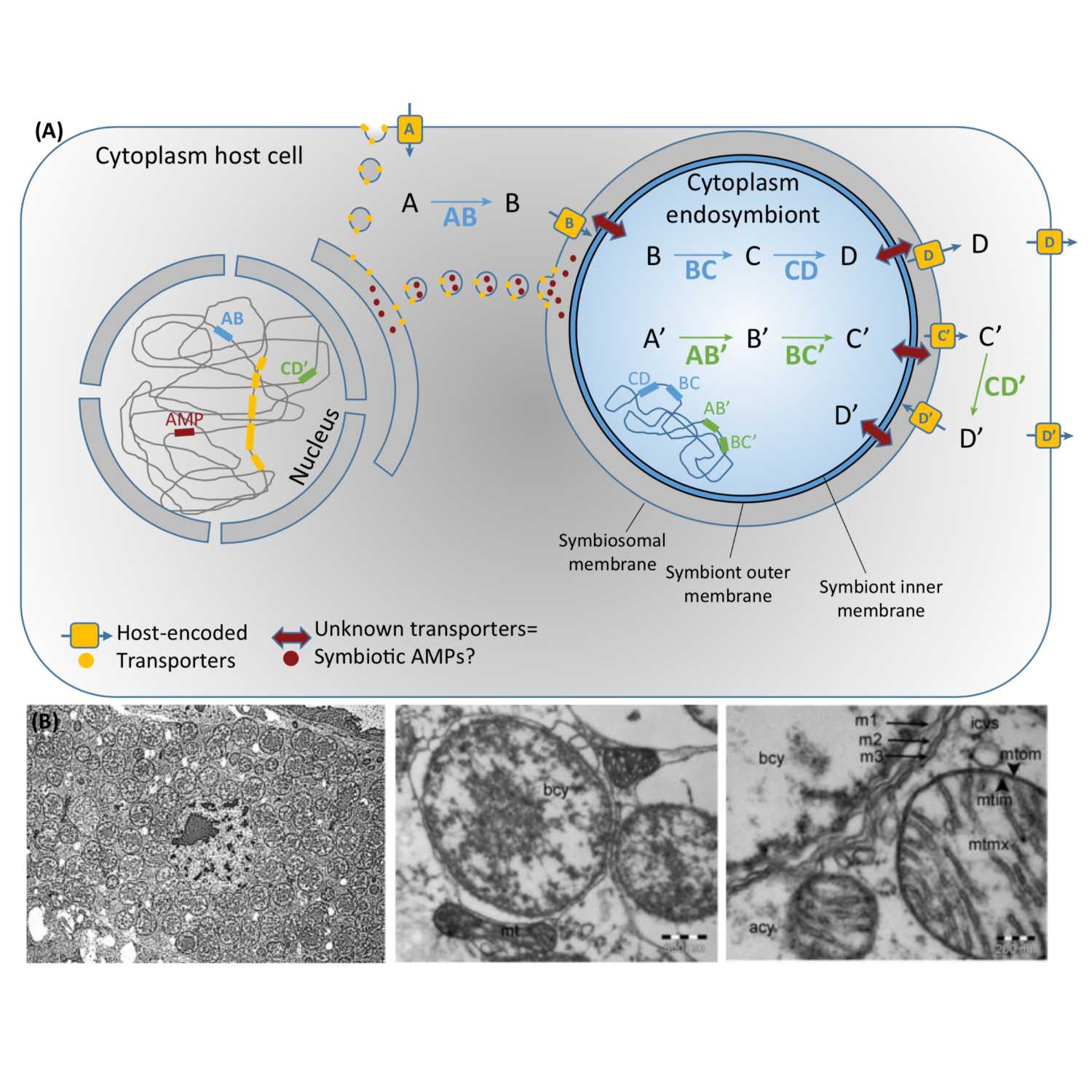
Papers Aloud#8 - Metabolic Integration of Bacterial Endosymbionts - Mergaert et al., 2017 - Cell PressTitle: Metabolic Integration of Bacterial Endosymbionts through Antimicrobial PeptidesAuthors: Peter Mergaert, Yoshitomo Kikuchi, Shuji Shigenobu, and Eva C.M. NowackCitation: Mergaert, P. et al. (2017) Metabolic Integration of Bacterial Endosymbionts through Antimicrobial Peptides. Trends Microbiol., 25, 703–712.Link to article
2021-04-3030 min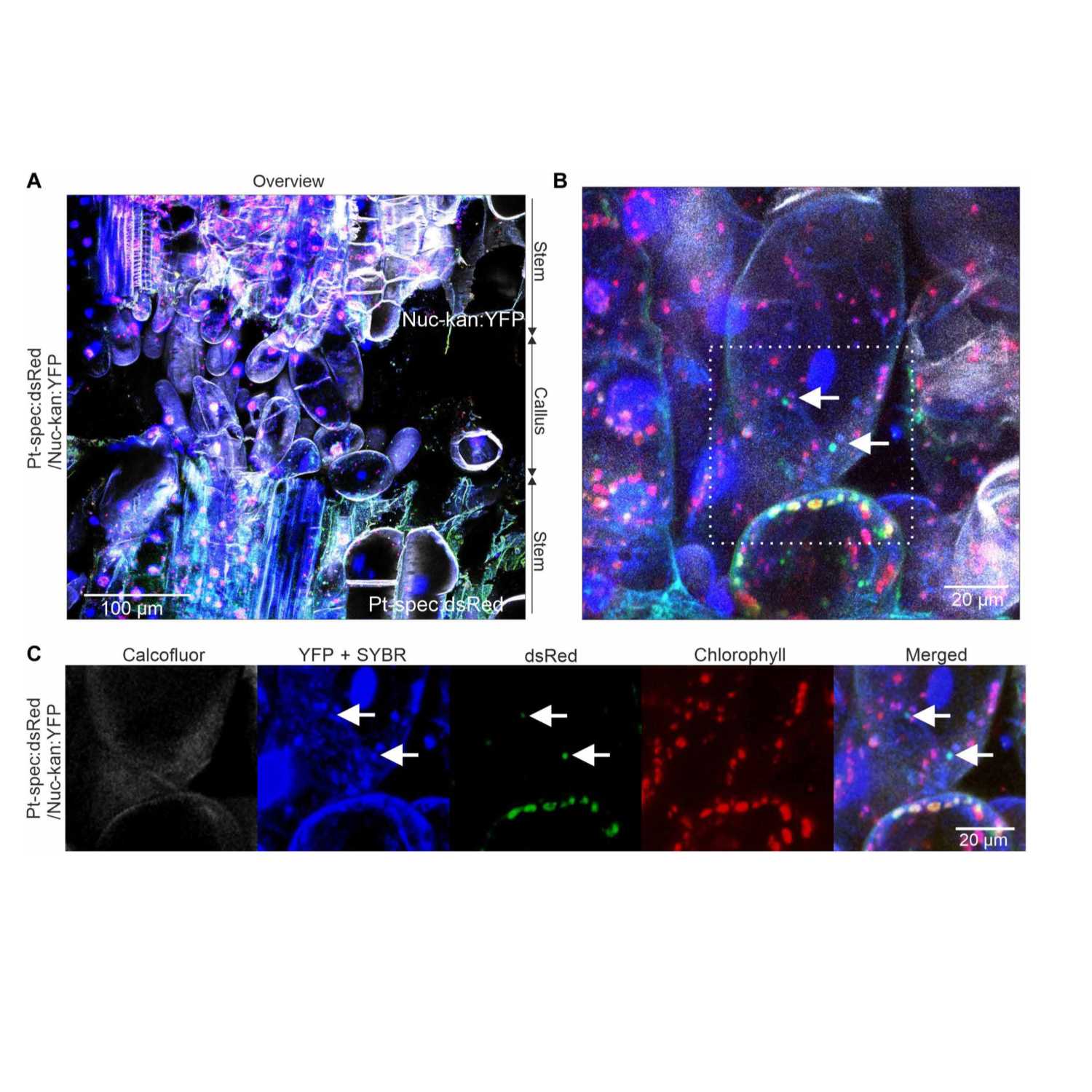
Papers Aloud#7 - Horizontal genome transfer - Hertle et al., 2021 - Science AdvancesTitle: Horizontal genome transfer by cell-to-cell travel of whole organelles.Authors: Alexander P. Hertle, Benedikt Haberl, Ralph Bock.Citation: Hertle, A.P. et al. (2021) Horizontal genome transfer by cell-to-cell travel of whole organelles. Science Advances, 7, eabd8215.Link to Article: https://advances.sciencemag.org/content/7/1/eabd8215
2021-04-2711 min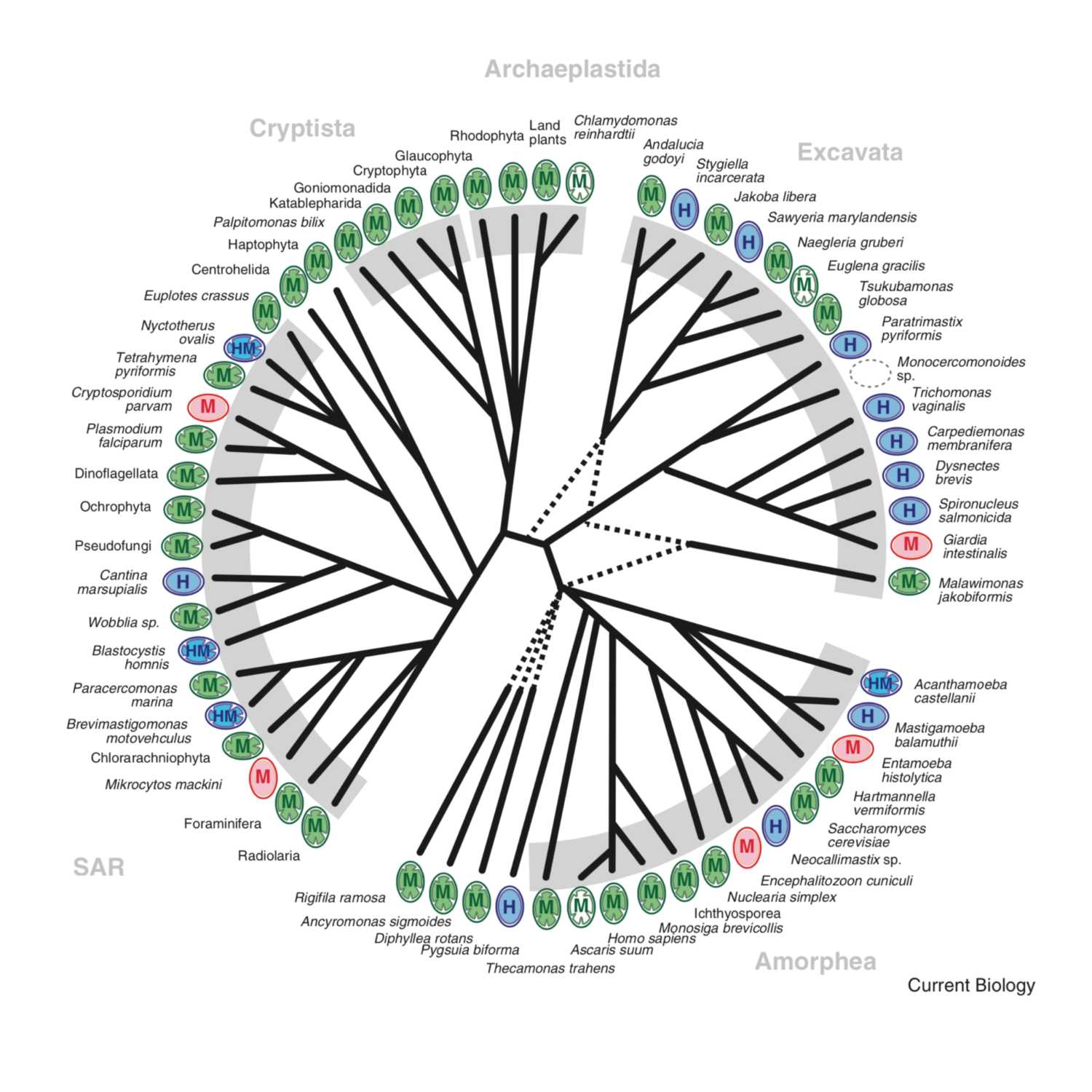
Papers Aloud#6 - Origin and Diversification of Mitochondria - Roger et al., 2017 - Current Biology ReviewTitle: The Origin and Diversification of Mitochondria. Authors: Andrew J. Roger, Sergio A. Muñoz-Gómez, and Ryoma Kamikawa. Citation: Roger,A.J. et al. (2017) The Origin and Diversification of Mitochondria. Curr. Biol., 27, R1177–R1192. Link to article: https://www.cell.com/action/showPdf?pii=S0960-9822%2817%2931179-X
2021-04-261h 15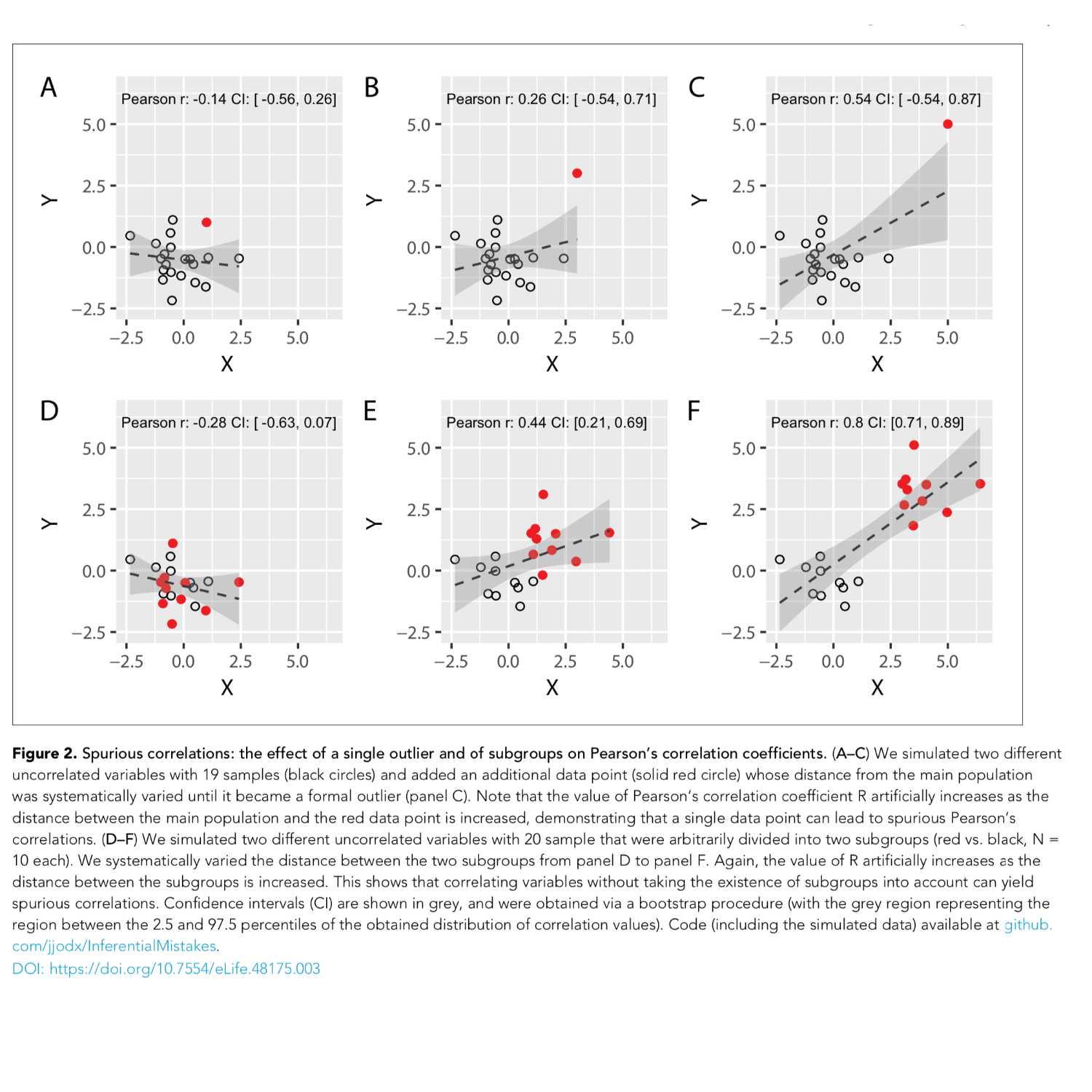
Papers Aloud#5 - Common Statistical Mistakes - Markin and Orban de Xivry, 2019Title: Ten common statistical mistakes to watch out for when writing or reviewing a manuscript.Authors: Tamar R. Markin and Jean-Jacques Orban de Xivry.Citation: Makin,T.R. and Orban de Xivry,J.-J. (2019) Ten common statistical mistakes to watch out for when writing or reviewing a manuscript. Elife, 8, e48175.Link to article: https://elifesciences.org/articles/48175
2021-04-2237 min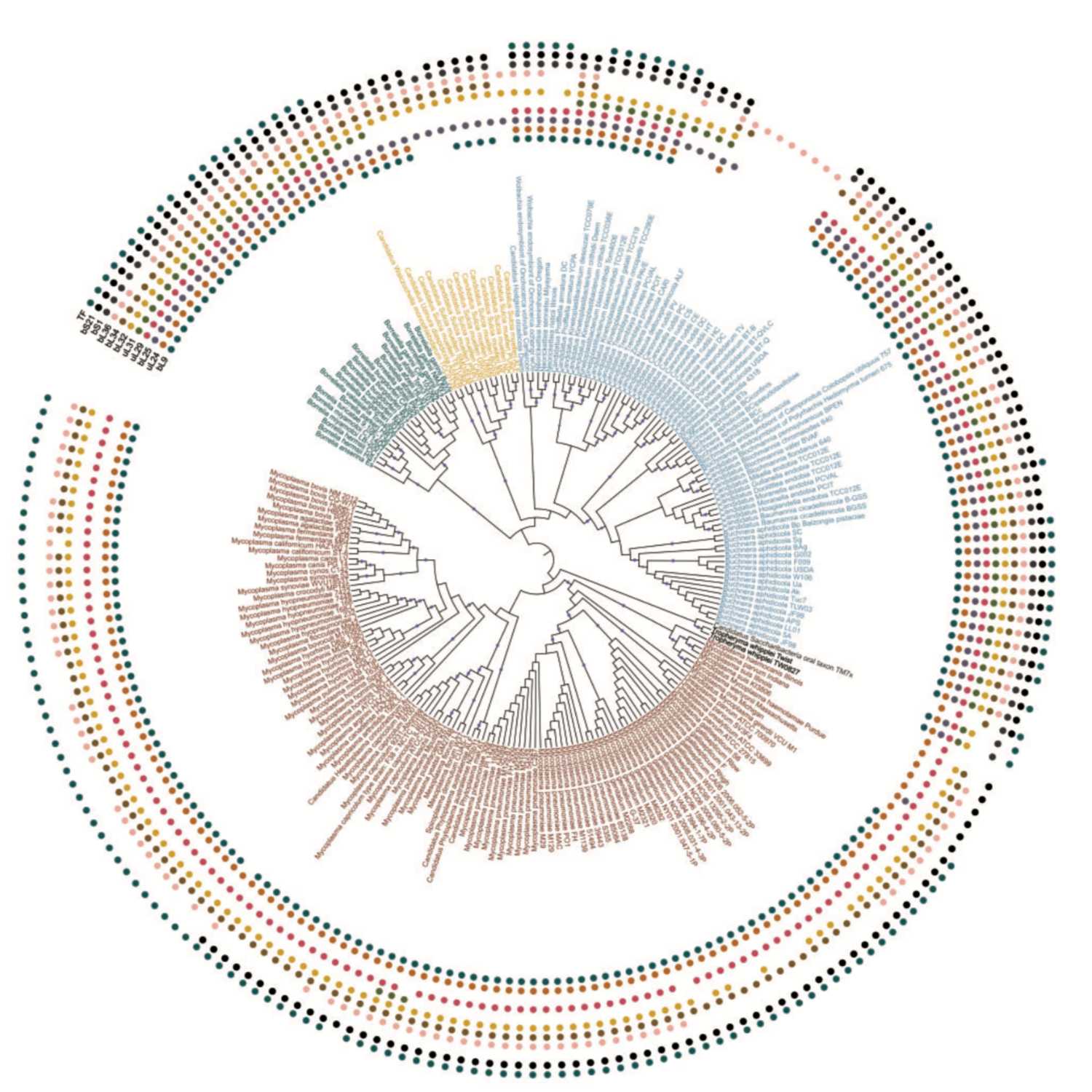
Papers Aloud#4 - Simplification of Ribosomes - Nikolaeva et al., 2021 - MBETitle: Simplification of Ribosomes in Bacteria with Tiny Genomes.Authors: Daria D. Nikolaeva, Mikhail S. Gelfand, and Sofya K. Garushyants.Citation: Nikolaeva, D.D. et al. (2021) Simplification of Ribosomes in Bacteria with Tiny Genomes. Mol. Biol. Evol., 38, 58–66. Link: https://academic.oup.com/mbe/article/38/1/58/5873528
2021-04-1919 min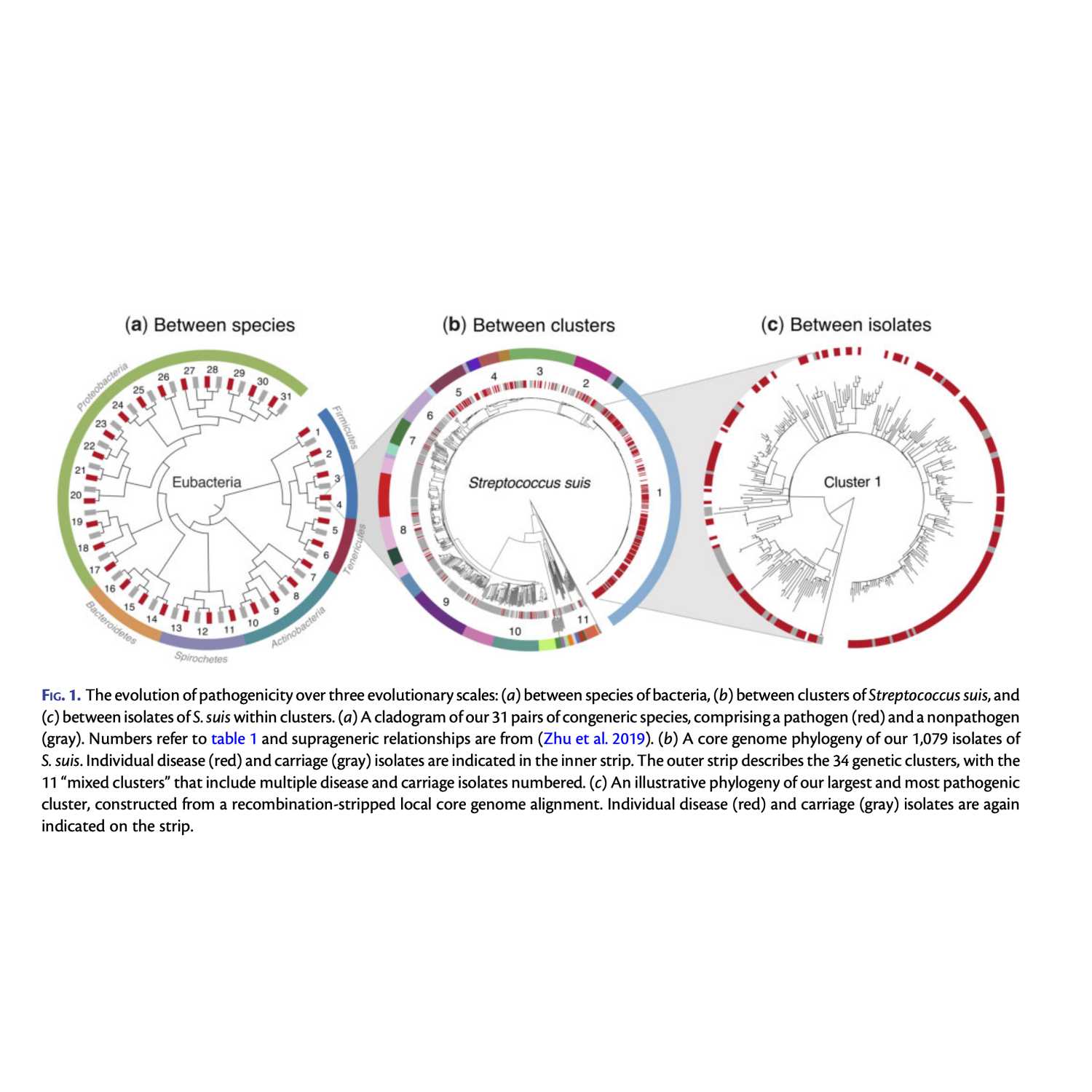
Papers Aloud#3 - Genome Reduction and Pathogenicity - Murray et al., 2020 - MBETitle: Genome Reduction Is Associated with Bacterial Pathogenicity across Different Scales of Temporal and Ecological Divergence.Authors: Gemma G.R. Murray, Jane Charlesworth, Eric L. Miller, Michael J. Casey, Catrin T. Lloyd, Marcelo Gottschalk, Alexander W. (Dan) Tucker, John J. Welch, and Lucy A. Weinert.Citation: Murray, G.G.R. et al. (2021) Genome Reduction Is Associated with Bacterial Pathogenicity across Different Scales of Temporal and Ecological Divergence. Mol. Biol. Evol., 38, 1570–1579. Link: https://academic.oup.com/mbe/article/38/4/1570/6031917
2021-04-1908 min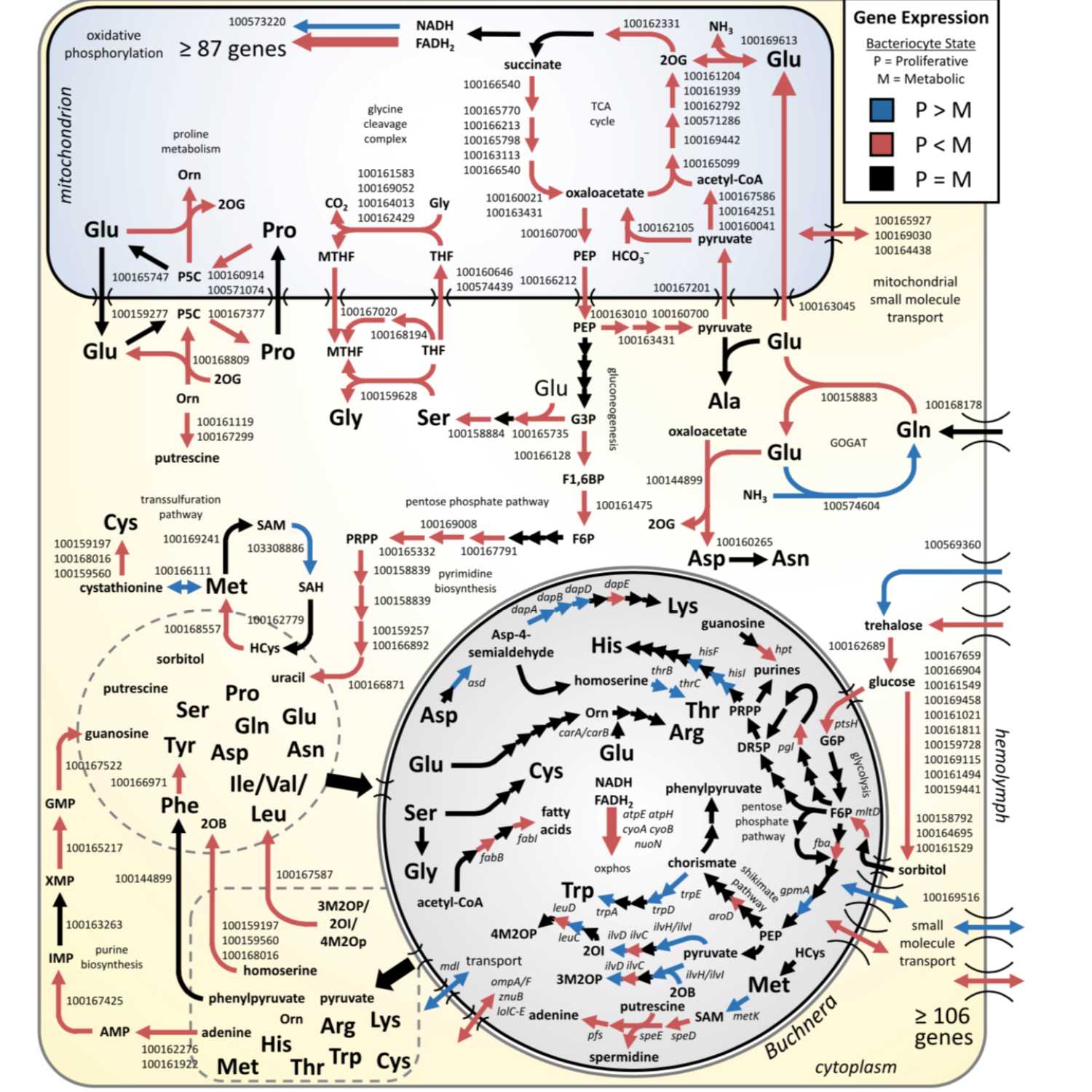
Papers Aloud#2 - Aphid-Buchnera Symbiosis - Smith and Moran, 2019 - PNASTitle: Coordination of host and symbiont gene expression reveals a metabolic tug-of-war between aphids and BuchneraAuthors: Thomas E. Smith and Nancy A. MoranCitation: Smith, T.E. and Moran,N.A. (2020) Coordination of host and symbiont gene expression reveals a metabolic tug-of-war between aphids and Buchnera. Proc. Natl. Acad. Sci. U. S. A., 117, 2113–2121. Link to article: https://www.pnas.org/content/117/4/2113
2021-04-1713 min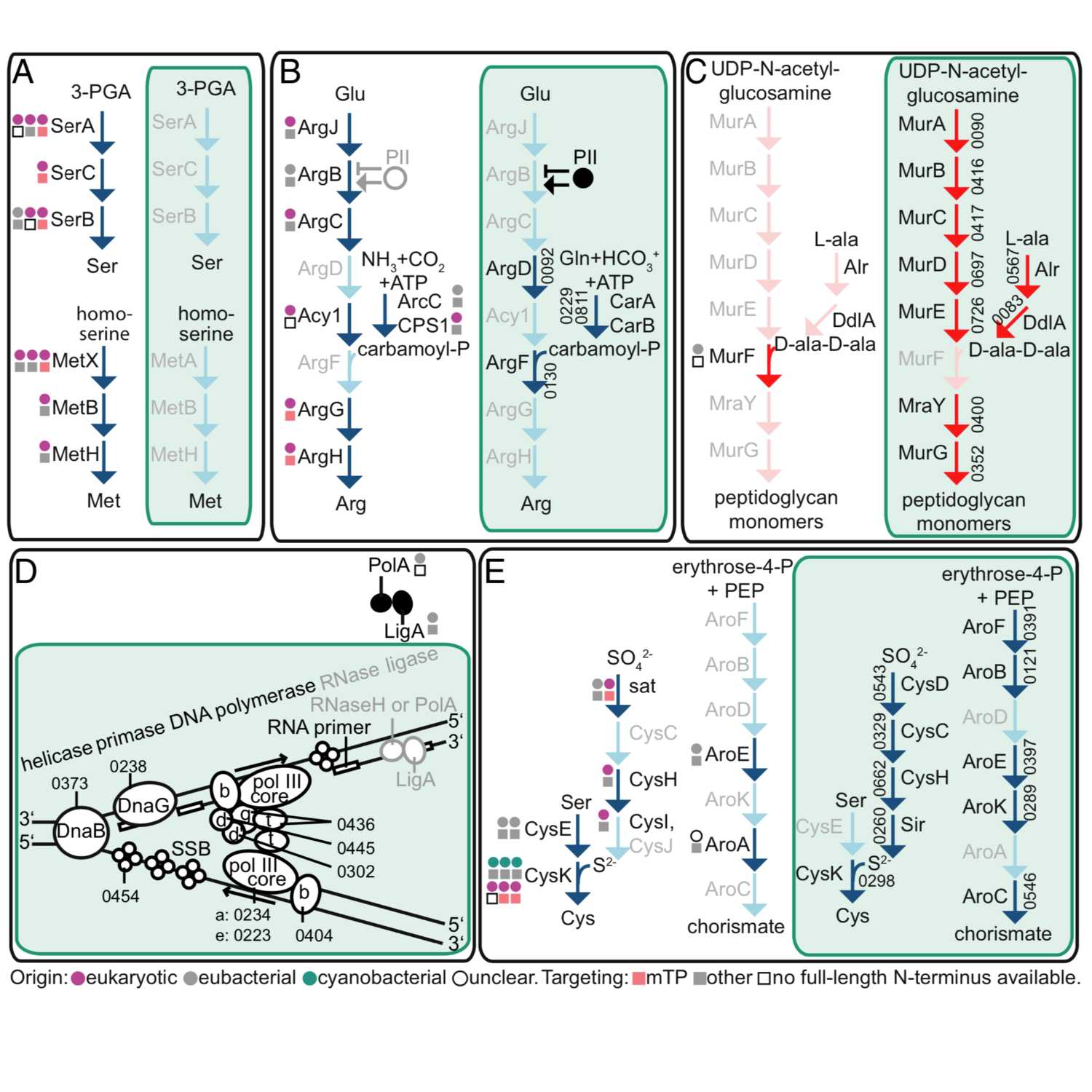
Papers Aloud#1 - Paulinella chromatophora symbiosis - Nowack et al., 2016 - PNASTitle: Gene transfers from diverse bacteria compensate for reductive genome evolution in the chromatophore of Paulinella chromatophora.Authors: Eva C. M. Nowacka, Dana C. Price, Debashish Bhattacharya, Anna Singer, Michael Melkonian, and Arthur R. Grossman.Citation: Nowack, E.C.M. et al. (2016) Gene transfers from diverse bacteria compensate for reductive genome evolution in the chromatophore of Paulinella chromatophora. Proc. Natl. Acad. Sci. U. S. A., 113, 12214–12219. Link to article: https://www.pnas.org/content/113/43/12214
2021-04-1529 min